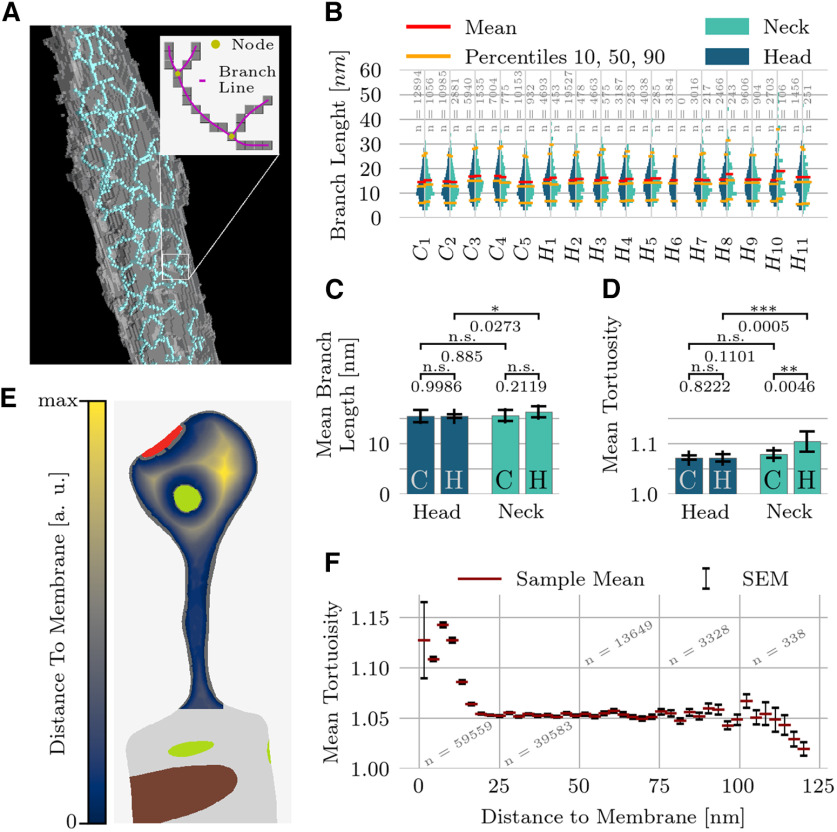
Figure 3.
Length and bending of branches. A, The topological skeleton is a set of connected voxels. A voxel connected to exactly two other voxels was considered to be part of a branch. A voxel connected to more than two other voxels was classified as a node (branching point). Voxels connected to only one other voxel were treated as branch ends. The branch length is computed as the arc-length of a smoothed curve through the voxels between two branching points (Materials and Methods). The tortuosity of a branch is defined as the ratio of the branch length to the Euclidean distance between the branch end points. B, Branch-length distributions of the entire spine population, shown separately for the head and neck regions. Sample size shown in gray. C, Pairwise comparison of mean branch length across hippocampal and cerebellar spines. Bar plots show the mean and STD of average branch lengths. D, Pairwise comparison of mean tortuosity. Bar plots show mean and STD of average branch tortuosity. E, Illustration of the normalized distance transform (DT), used here to measure the distance of any point within a spine to the spine membranes, which include the interfaces with the postsynaptic density and endoplasmic reticulum. F, Tortuosity is increased close to the membrane and constant elsewhere. As hippocampal neck volumes are smaller and therefore have a higher surface/volume ratio this can explain the differences in D.