Figure 2.
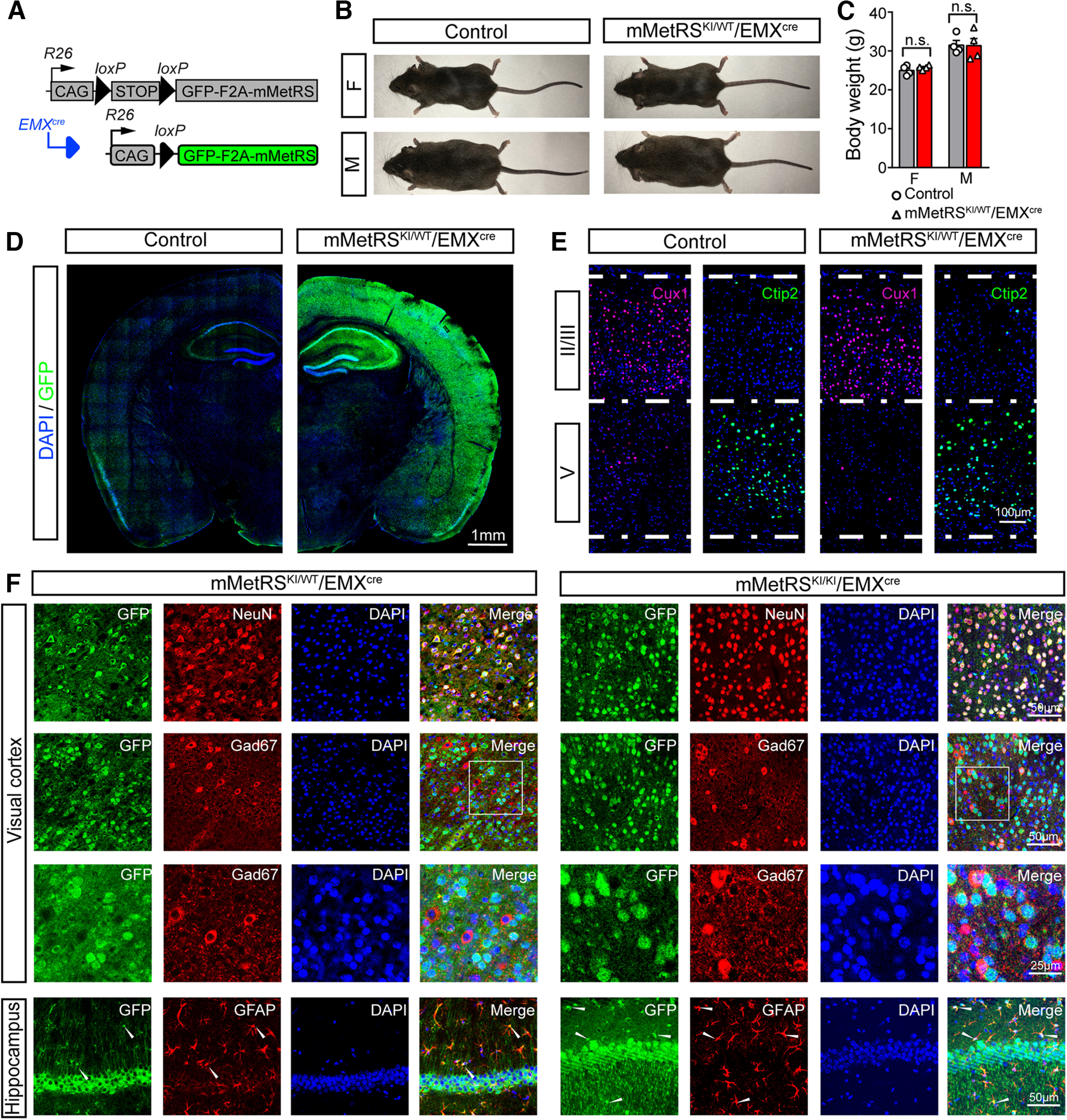
Characterization of mMetRS/EMXcre lines. A, Schematic of the breeding strategy. EMXcre drives expression of GFP-F2A-mMetRS from the Rosa26floxSTOP-GFP-F2A-mMetRS allele, allowing GFP expression to report mMetRS distribution. B, Images of age-matched animals of mMetRS/EMXcre mutants and their MetRS-negative control littermates at postnatal day 60. C, Body weights of male and female mMetRS/EMXcre mutants and control littermates are comparable (individual datapoints and mean ± SEM values; N = 4, paired t test with Welsh's correction, n.s., not significant). D, GFP expression in cortex and hippocampus in coronal brain sections counterstained with DAPI. E, Confocal images of immunolabeling of layer-specific excitatory cortical neurons with antibodies to Cux1 and Ctip2 in layers II/III and V in DAPI-stained coronal sections in visual cortex, indicating that mMetRS expression does not affect the development of cortical lamination. F, Cellular specificity of GFP-F2A-mMetRS expression in mMetRSKI/WT/EMXcre and mMetRSKI/KI/EMXcre mice. Coronal sections through visual cortex and hippocampus were labeled with DAPI and antibodies against markers of all neurons (NeuN), inhibitory GABAergic neurons (GAD67), or astrocytes (GFAP). GFP is present in NeuN+ neurons, but not in GABAergic neurons. Some GFAP+ astrocytes express GFP, which is consistent with the known EMX expression pattern.
