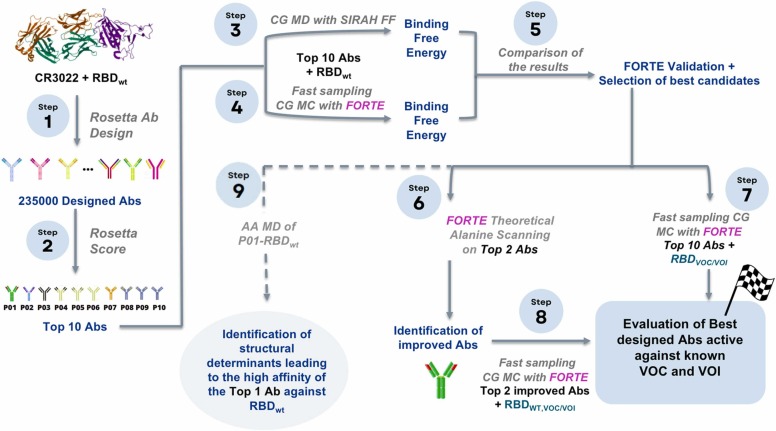
Fig. 1.
Scheme for the multiple scales in silico protocol, consisting of an initial structural-bioinformatics-based methodology to explore macromolecules as potential candidates (steps 1 and 2), constant protonation state CG MD (steps 3), constant-pH (variable protonation state) CG MC simulations (steps 4, 6–8), and an atomistic constant charge MD simulation (step 9). At the end of this cycle, an optimized mAb with a higher binding affinity is obtained. See the text and the SM for more details.