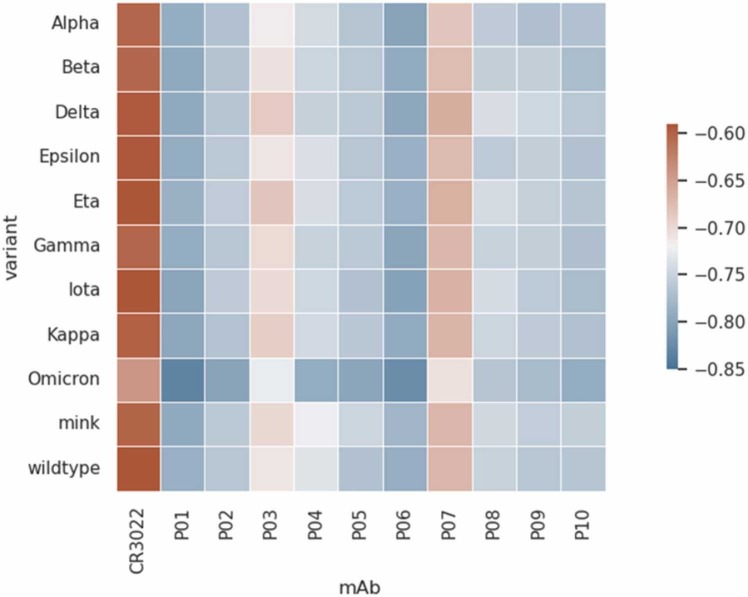
Fig. 6.
Heatmap with the minima free energy of interactions values (βwmin in KBT units) for the SARS-CoV-2 RBD-mAbs complexation at pH 7 and 150 mM of NaCl by the CpH MC simulations (FORTE) for the RBD of the main critical variants and different Rosetta-designer binder candidates (P01 to P10). The reference is the native fragment of CR3022. All values of. The maximum estimate error is 0.01. See the text for a description of mutations considered in each case.