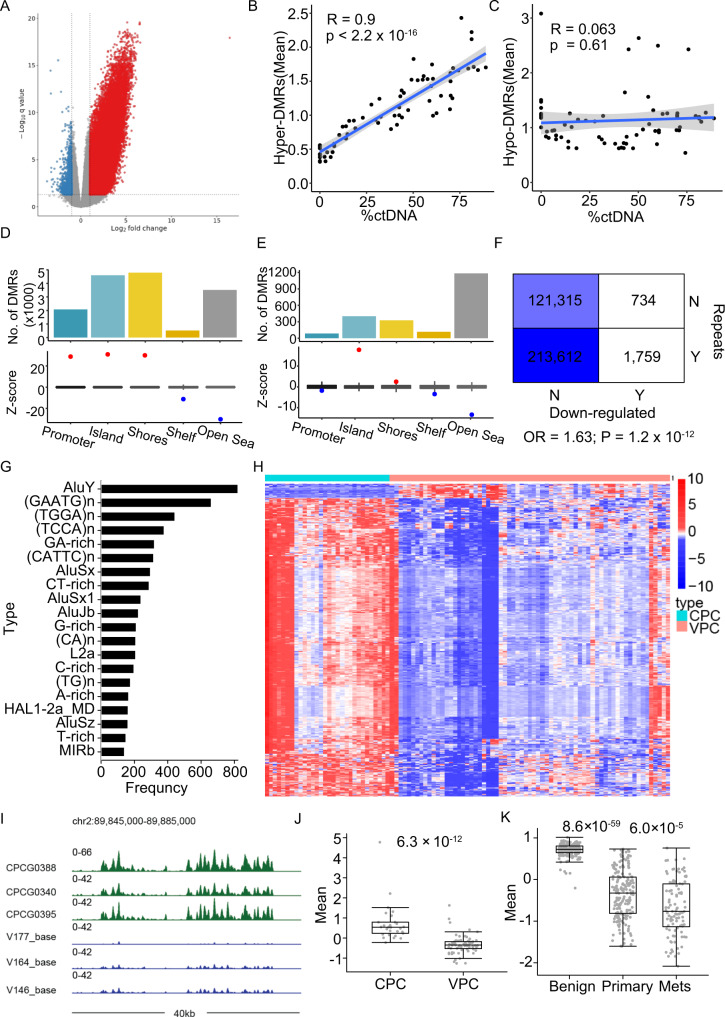
Fig. 2. cfMeDIP reveals widespread hypermethylation and preferential repeat hypomethylation in metastatic samples.
A Volcano plot of differentially methylated regions (DMRs) was identified comparing metastatic and localized prostate cancer. A total of 2493 and 19,048 hypo and hyper DMRs were identified, respectively. Pearson correlation between the mean methylation levels of hyper (B) and hypo (C) DMRs and %ctDNA. P value was calculated using t test. Blue line represents a fitted linear model of the data and shading around the fitted line represents 0.95 CI. Genomic distribution of hyper- (D) and hypo- (E) DMRs. F Contingency table showing the distribution of repeat and the hypo-DMRs. Two-sided fisher’s exact test was used to calculate p value and odds ratio (OR). G Frequency of repeat types overlapped with downregulated peaks. H Differentially methylated peaks located within the 1 Mb regions flanking the centromere. CPC Canadian Prostate Cancer Genome Network cohort. I Example showing signal distribution around the pericentromeric region in chromosome 2. The Y axis showed the normalized signal per million reads (SPMR) from MACS (v2.2.5). Mean methylation levels for differential peaks shown in H for cfMeDIP-seq (J) and WGBS (K) data. Box plots represent median values and 0.25 and 0.75 quantiles. Whiskers represent 1.5× IQR. X = 30, and 67 independent experiments for the CPC and VPC cohorts, respectively in J. X = 194, 194, and 100 independent experiments for the Benign, Primary, and Mets groups, respectively, in K. Source data for F2B-C and 2J-K are provided as a Source Data file.