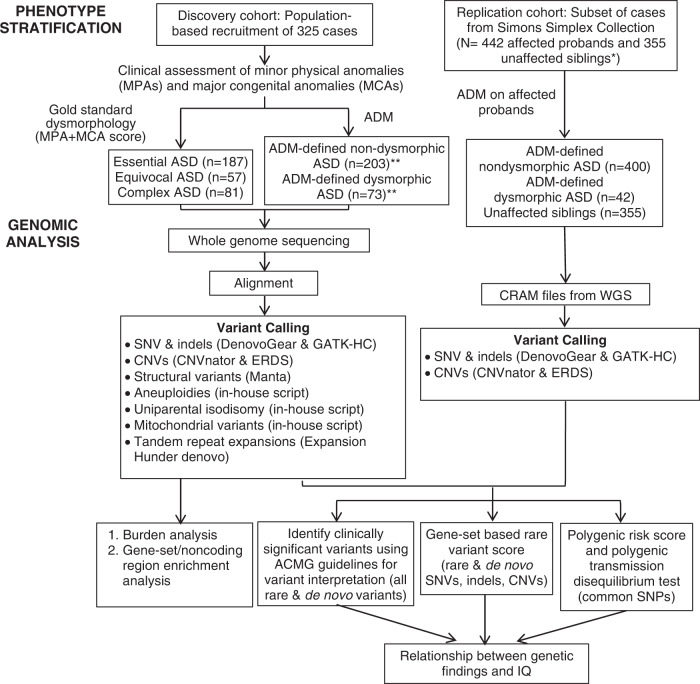
Fig. 1. Project workflow.
Summary of phenotype stratification, whole-genome sequencing workflow, and genomic analyses performed in this study. ASD autism spectrum disorder, ADM autism dysmorphology measure, CNVs copy-number variants, SNVs single-nucleotide variants, indels insertions and deletions, ERDS estimation by read depth with single-nucleotide variants, GATK-HC Genome Analysis Toolkit-Haplotype Caller, SNPs single-nucleotide polymorphisms, ACMG American College of Medical Genetics and Genomics, IQ intelligence quotient. *Unaffected siblings were used for GRVS and PRS analyses. **Excluding samples with false negative ADM-defined nondysmorphic ASD. We also included only samples sequenced on Illumina platforms to be consistent with the replication cohort. For variant calling, on average per sample, we detected ~3.7 million SNPs, 36,514 rare single-nucleotide variants (SNVs), 4113 small insertions and deletions (indels), 13 rare copy-number variants (CNVs), 390 rare SVs, 73.4 de novo SNVs, 7.3 de novo indels, and 0.1 de novo CNVs (Supplementary Data 2). Experimental validation rates were 94.8%, 85.7%, and 87.5%, respectively, for de novo SNVs, indels, and CNVs (Supplementary Data 3 and 4). Using GRVS, we were able to quantify and validate the contribution of morphology-associated, rare sequence-level and copy-number variants to morphological ASD subtypes. While we can call other SVs from the WGS, there needs to be higher-quality data before these can be effectively incorporated into GRVS.