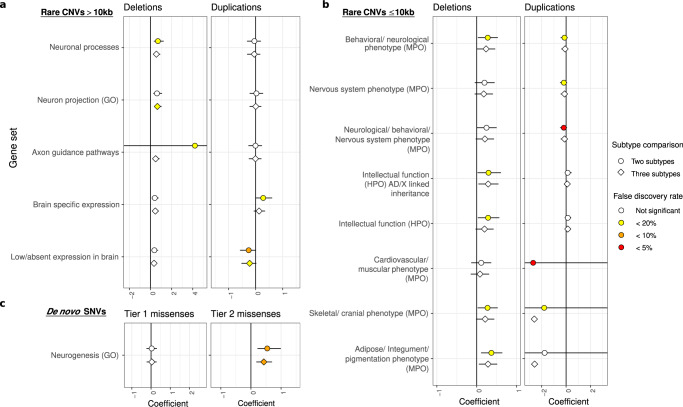
Fig. 2. Gene sets for which de novo and rare coding variants are significantly more prevalent in some subtypes of ASD.
We define events as (a, b) genes impacted by CNVs or (c) as variants for SNVs and indels. The coefficient is the relationship between the number of events in each gene set and the ASD subtypes; it reflects the effect size of a variant type and gene set among different ASD subtypes. Positive coefficients indicate more events in individuals with ASD and more dysmorphic features; negative coefficients indicate more events in individuals with ASD and fewer dysmorphic features. We show only gene sets for which (a, b) rare CNVs (n = 325 samples), or (c) de novo missense variants (n = 235 samples) are significantly more prevalent in different subtypes of ASD. Tier 1 and 2 missense variants consist of all or only predicted damaging missense variants, respectively, as defined in ref. 25. Symbol shapes indicate the subtype comparisons that were conducted for each combination of the gene set and variant type. Two-sided likelihood ratio tests were performed with permutation-based FDR for the multiple-testing correction. Two subtype comparison = nondysmorphic vs. dysmorphic ASD. Three subtype comparison = essential vs. equivocal vs. complex ASD. Colored shapes indicate significant signals after multiple test correction by permutation-based FDR, where yellow, orange, and red, indicate permutation-based FDR < 20%, 10% and 5%, respectively. The data points (the center) indicate the estimated coefficient, while error bars indicate 95% confidence intervals of the estimated coefficient. Source data are provided as a Source Data file.