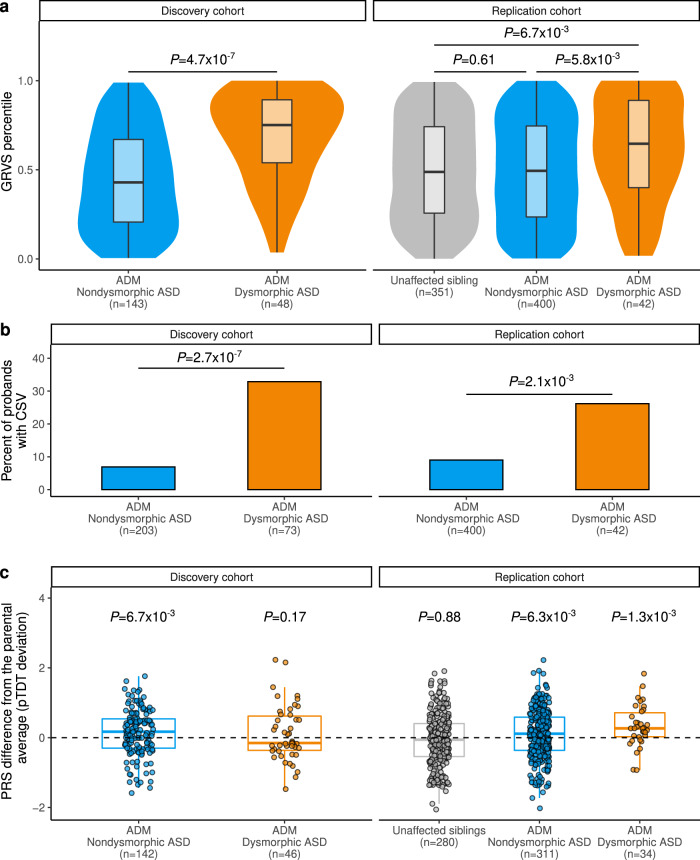
Fig. 6. Replication of rare and common genetic findings in a subset of Simons Simplex Collection cohort.
a GRVSs for each cohort were ordered and ranked by percentile. Violin plots show the distributions of the probands’ GRVS percentiles; box plots contained within show the median and quartiles of the percentiles for each subtype, the minima and maxima of box plots indicate 3× the interquartile range-deviated scores from the median. P values denote the probability that the GRVS in ADM-defined dysmorphic ASD is not greater than ADM-defined nondysmorphic ASD (one-sided, Wilcoxon rank-sum test). b Yield of CSVs between dysmorphic and nondysmorphic subtypes in discovery and replication cohorts. P values indicate the probability that the yield of CSVs in nondysmorphic ASD is not lower than that of dysmorphic ASD (one-sided, Welch’s t-test). c Inheritance of polygenic risk for ASD in dysmorphic and nondysmorphic ASD subtypes in discovery and replication cohorts. Box plots depict the median and quartiles of pTDT deviation, and the minima and maxima of box plots indicate 3× the interquartile range-deviated pTDT deviations from the median. Dots represent pTDT deviations of subjects. P values for each subgroup indicate the probability that the mean of the pTDT deviation distribution is not greater than zero (one-sided, Welch’s t-test), as depicted by the dotted line. The finding of no significant overtransmission in dysmorphic ASD did not replicate in SSC, which might be due to lack of statistical power (i.e., at least 100 dysmorphic samples are needed to achieve 80% power if PRS explains 2.45% of phenotypic variance11) and/or ascertainment differences between the discovery and replication cohorts. The discovery cohort included data about major congenital anomalies in morphologic classification, whereas the replication cohort did not. While our discovery cohort was population-based, the Simons Simplex Collection excluded probands with medically significant perinatal diseases, severe neurological deficits, and certain genetic syndromes32. This likely decreased the proportion of probands with excess MPAs and birth defects, potentially leading to a lower burden of common ASD-associated variants88. Source data are provided as a Source Data file.