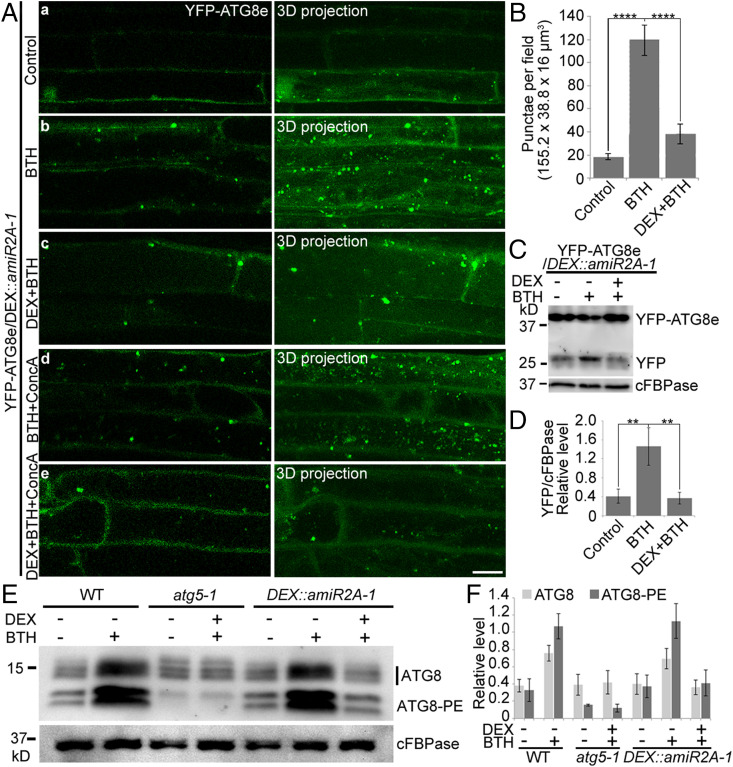
Fig. 5.
ORP2A is required for plant autophagy. (A) KD of ORP2A affected autophagy. Seven-day-old YFP-ATG8e/DEX::amiR2A seedlings were treated with BTH (b), DEX+BTH (c), BTH+ConcA (d), and DEX+BTH+ConcA (e) for 6 h, followed by confocal imaging and 3D projection. ΔZ = 0.8 μm, 20 stacks. For DEX treatments, DEX was pretreated for 48 h prior to other treatments. (Scale bar, 10 μm.) (B) Quantification of YFP-ATG8e–positive punctae per root section in A. Error bars indicate ±SD; ****P ≤ 0.0001, Student’s t test. (C) Immunoblot detection of YFP-ATG8e turnover in transgenic YFP-ATG8e/DEX::amiR2A-1 plants. Seven-day-old seedlings of YFP-ATG8e/DEX::amiR2A-1 were subjected to treatments of DEX/BTH/ConcA as indicated, followed by protein extraction and immunoblot detection with GFP antibodies. cFBPase antibodies were used as a loading control. PE, phosphatidylethanolamine. (D) Quantification of free YFP normalized to cFBPase in C. At least three independent replicates; error bars indicate ±SD; **P ≤ 0.01, Student’s t test. (E) Immunoblot detection of the ATG8 lipidation level in WT, atg5-1, and DEX::amiR2A-1. Seven-day-old seedlings of WT, atg5-1, and DEX::amiR2A-1 were treated with/without BTH for 6 h and/or DEX for 48 h as indicated, followed by protein extraction and immunoblot analysis with ATG8 antibodies. cFBPase antibodies were used as a loading control. (F) Quantification of ATG8 upper (ATG8) and lower (ATG8-PE) bands normalized to cFBPase in E. At least three independent replicates; error bars indicate ±SD.