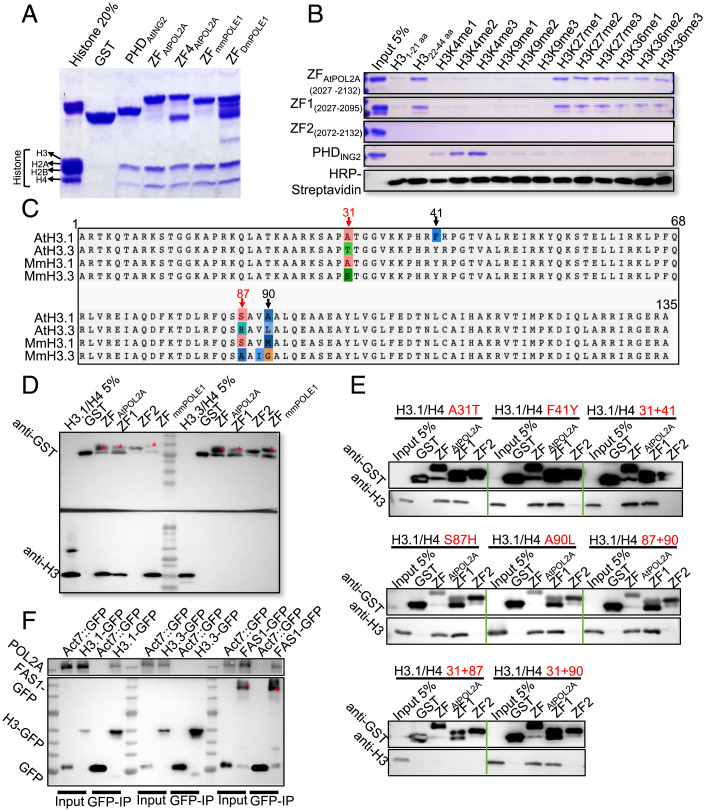
Fig. 6.
ZF1 rather than ZF2 of POL2A binds to H3.1 both in vitro and in vivo. (A) Calf histone pull-down assays of glutathione S-transferase (GST)–fused ZFs of POL2A show that ZFPOL2A mainly binds to H3; the PHD of AtING2 is used as a positive control. ZFPOL2A (2027-2125) and ZF4POL2A (2034–2125) are truncated proteins harboring POL2A ZF1 and ZF2. (B) Biotinylated histone peptide pull-down assays of GST-fused ZFs, with AtING2 Plant Homeodomain (PHD) as a positive control. The biotin-labeled peptides were detected via Horseradish Peroxidase (HRP)-Streptavidin. (C) Alignment of H3.1 and H3.3 orthologs between Arabidopsis and mouse with four-residue difference marked above. (D) GST pull-down assay showing ZF-binding activity to H3.1/H4 and H3.3/H4 dimers. Red stars indicate the band corresponding to the predicted size of each protein. (E) GST pull-down assay showing ZF-binding activity to different mutated H3.1/H4 proteins corresponding to C. Red texts indicate the types of point mutation of H3.1. (F) POL2A coimmunoprecipitated with H3.1-GFP (HTR13::HTR13-GFP) and FAS1-GFP (Act7::FAS1-GFP), rather than H3.3-GFP (HTR5::HTR5-GFP), with Col-0 plants expressing empty GFP-tag vector as negative control. Red stars indicate the band corresponding to the predicted size of FAS1-GFP.