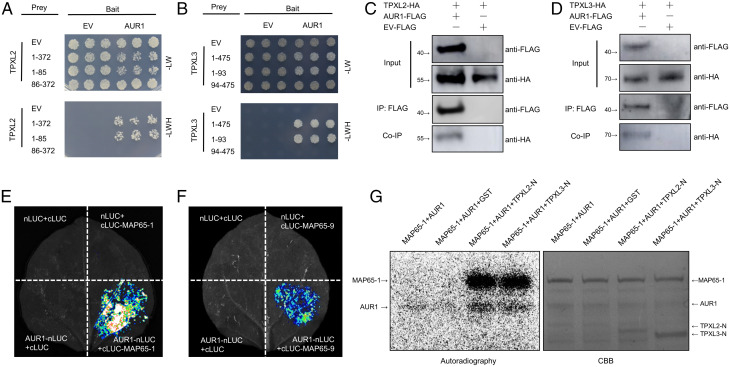
Fig. 2.
AUR1 interacts with multiple microtubule-associated proteins. (A) Y2H assays between AUR1 and TPXL2. The combinations of proteins expressed in either the prey vector (pGADT7) or the bait vector (pGBKT7) are indicated alongside the yeast colonies. The full-length of TPXL2 is indicated by 1-372, 1-85 indicates the N terminus of TPXL2, 86-372 indicates the C terminus of TPXL2. Yeast cells were plated onto SD-2/-Leu-Trp medium and SD-3/-Leu-Trp-His medium. (B) Y2H assays to test for interaction between AUR1 and TPXL3. The full-length of TPXL3 is indicated by 1-475, 1-93 indicates the N terminus of TPXL3, 94-475 indicates the C terminus of TPXL3. (C and D) Co-IP assays of AUR1-FLAG and TPXL2-HA (C) or TPXL3-HA (D) in N. benthamiana leaves. Proteins were immunoprecipitated (IP) with anti-FLAG-M2 beads and analyzed by Western blot using horseradish peroxidase–conjugated anti-FLAG or anti-HA antibody. (E and F) Split luciferase (LUC) complementation assays between AUR1 and MAP65-1 (E) or MAP65-9 (F). nLUC (N terminus of LUC)-tagged AUR1 was coinfiltrated into N. benthamiana leaves along with the cLUC (C terminus of LUC)-tagged MAP65-1 or MAP65-9. (G) Phosphorylation of recombinant MAP65-1 protein by AUR1 in the presence of TPXL2-N or TPXL3-N. An in vitro phosphorylation reaction was performed using purified proteins and 32P-labeled ATP and detected by autoradiography. The Upper band (Left panel) shows the phosphorylation of MAP65-1 by AUR1, and the bottom band shows AUR1 auto-phosphorylation. CBB, Coomassie brilliant blue staining. Experiments were carried out two (G) or three times (A–F) with similar results. EV, empty vector; HA, hemagglutinin.