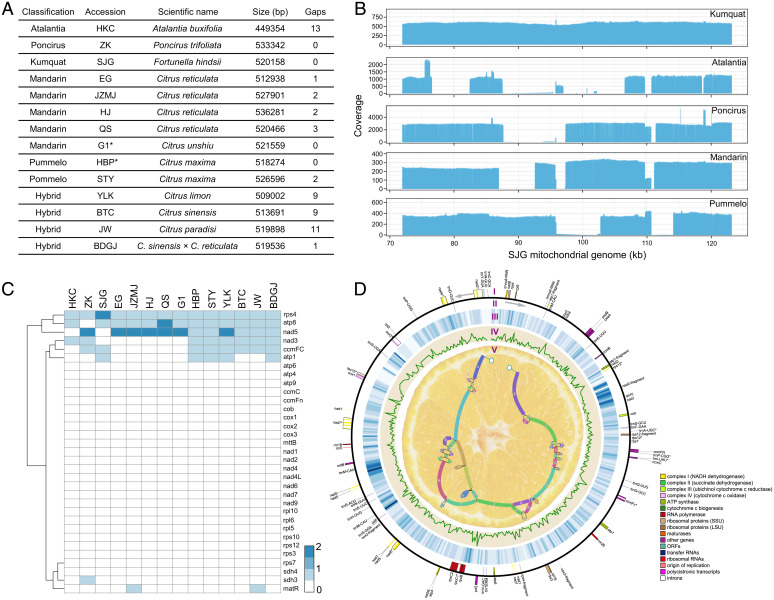
Fig. 1.
SVs and chimeric ORFs in the citrus super pan-mitogenome. (A) Summary of the 12 de novo mitochondrial assemblies and 2 previously published mitochondrial genomes. The previously published sequences are highlighted with asterisks. (B) 70- to 120-kb regions identified based on the coverage depth of long-read mapping containing >5-kb deletions from five groups. The mitogenomic regions are shown on the x-axis. The different levels of coverage are indicated on the y-axis. (C) Chimeric ORFs related to conserved genes in the intergenic regions of 14 citrus mitochondrial genomes. The heatmap includes two previously reported mitochondrial genomes, mandarin G1 and pummelo HBP. (D) Circular representation of the mitochondrial pan-genome. The conserved gene structure was annotated for the forward strand (I) and the reverse strand (II). The heatmap indicates the distribution of SNPs and indels among the 184 accessions of citrus, with the mitochondrial genome from kumquat used as a reference (III). The diversity of SNPs and indels in the mitochondrial genomes in 184 citrus accessions is indicated (IV). The mitochondrial pan-genome is indicated with a schematic drawing (V).