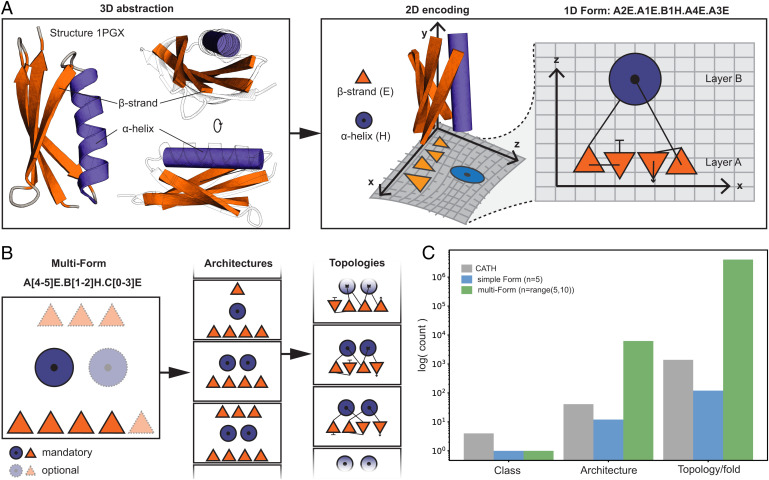
Fig. 1.
Form parametrization for protein structures. (A) Form parametrization for an ubiquitin-like fold (PDB 1PGX). A 3D abstraction of the structure is created that is encoded into a layered two-dimensional (2D) lattice diagram where the sheet is assigned as layer A and the helix as layer B (layer assignment is arbitrary). The SSEs on a layer are dispersed on the x axis, and layers are stacked onto each other following the z axis. The lattice representation is summarized into a form string as shown on the Top. The form describes each SSE by the layer, relative position in the layer, and secondary structure type separated by a dot (N- to C-terminal sequence order is preserved). (B) A multiform string created by assigning some SSEs as mandatory and others as optional. The flexibility allows the sampling of a range of architectures and topologies. (C) Comparing the exploratory capacity between a simple form (5 SSEs), a multiform (a minimum of 5 and a maximum of 10 SSEs) and the known space of protein folds (as classified by CATH). The simple form nearly samples as many existing topologies as known, while the multiform greatly generates more topologies than what can be found in nature.