Figure 5.
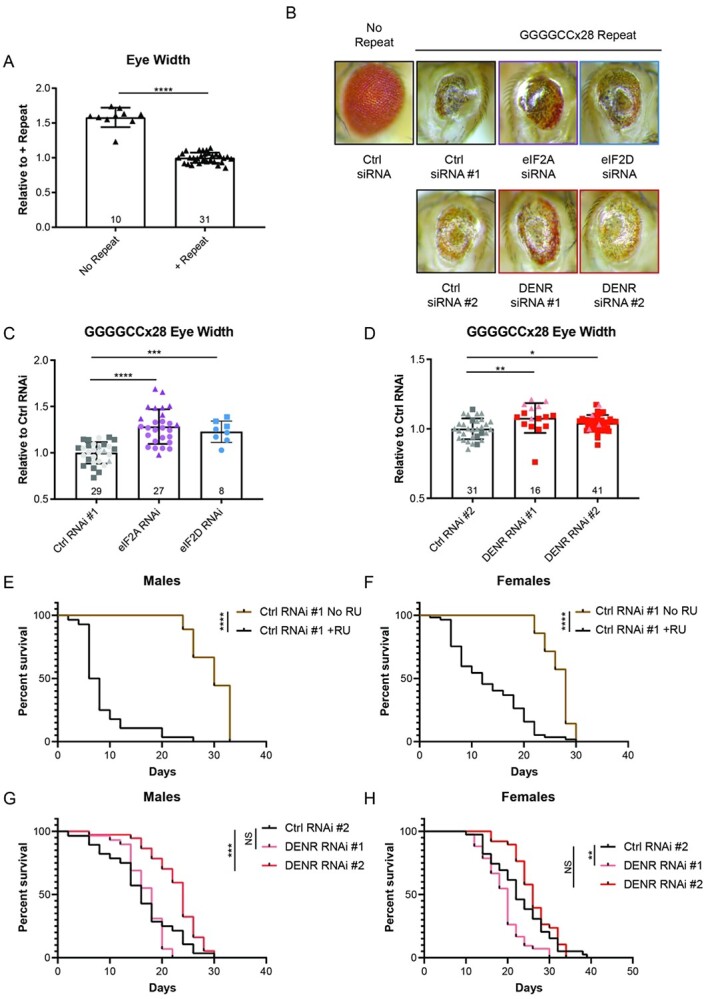
Knockdown of DENR suppresses GGGGCC associated toxicity in Drosophila. (A) Quantification of eye width in flies expressing control (ctrl) shRNA #1 in the absence or presence of the GGGGCCx28 repeat, under the GMR-Gal4 driver. Experimental numbers are indicated within bars and each fly is represented by a single data point. (B) Representative images of fly eyes expressing control or CG7414 (‘eIF2A’), eIF2D, or DENR shRNAs in the presence of the GGGGCCx28 repeat. (C–D) Comparison of eye width in flies expressing indicated shRNAs in the presence of the GGGGCCx28 repeat. Experimental numbers are indicated within bars, and each fly is represented by a single data point. Shape of data points represents the experimental trials that data points were collected from. (E–F) Survival curve of control male and female Tub5-GS-Gal4 flies, respectively. Survival of flies with induced expression of the GGGGCCx28 repeat and control shRNA through RU486 treatment (+RU, male n = 28, female n = 57), is compared to survival of flies without induced expression (No RU, male n = 9, female n = 14). (G–H) Survival curve of RU486-treated GGGGCCx28 male and female Tub5-GS-Gal4 flies, respectively, expressing ctrl (male n = 28, female n = 39) or DENR targeting shRNA #1 (male n = 29, female n = 42) and shRNA #2 (male n = 37, female n = 38). Bar graphs represent mean with error bars ± standard deviation, * P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by one-way ANOVA with Dunnett’s multiple comparison test. For survival curves, **P < 0.01, ***P < 0.001, ****P < 0.0001, Mantel-Cox test.
