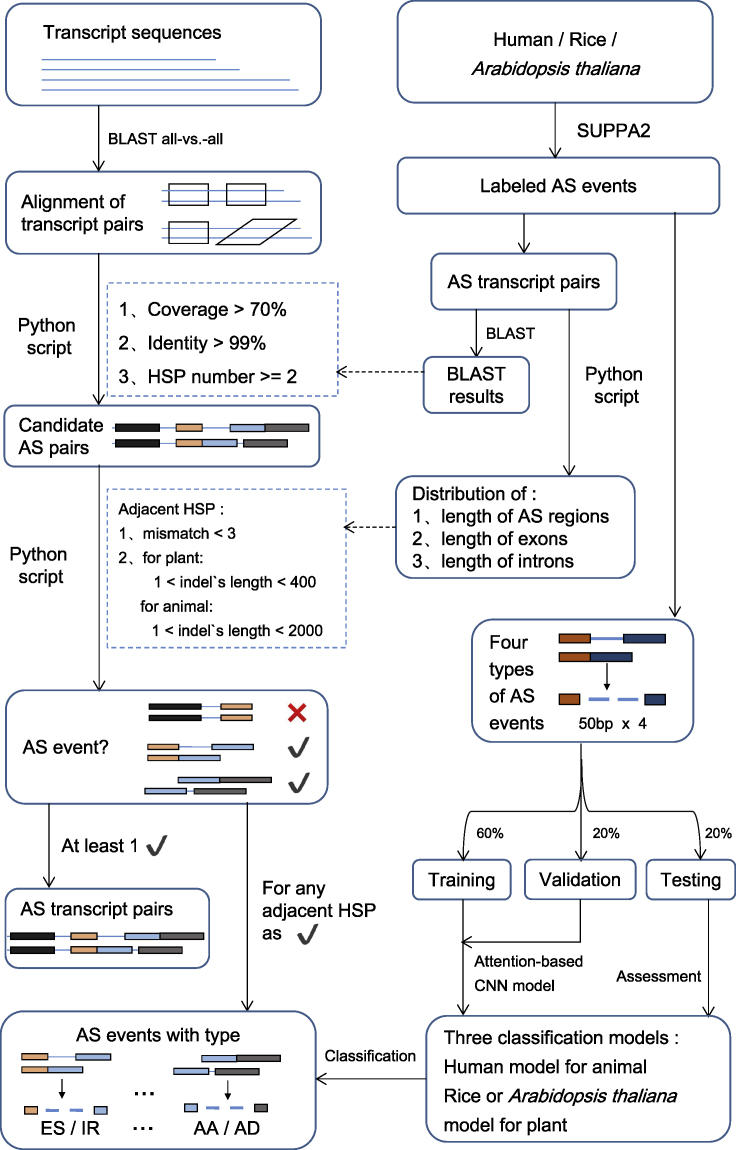
Figure 1.
The workflow of DeepASmRNA
DeepASmRNA is composed of two parts: identification of alternatively spliced transcripts and classification of AS events. For identification, identifying AS patterns was based on the results of all-versus-all alignment transcripts obtained by BLASTN. For classification, an attention-based CNN model was used to classify the AS events into 4 types. HSP: high-scoring segment pairs in BLAST, which indicated constitutive region. Indels: insertion and deletion, which indicated the alternative region.