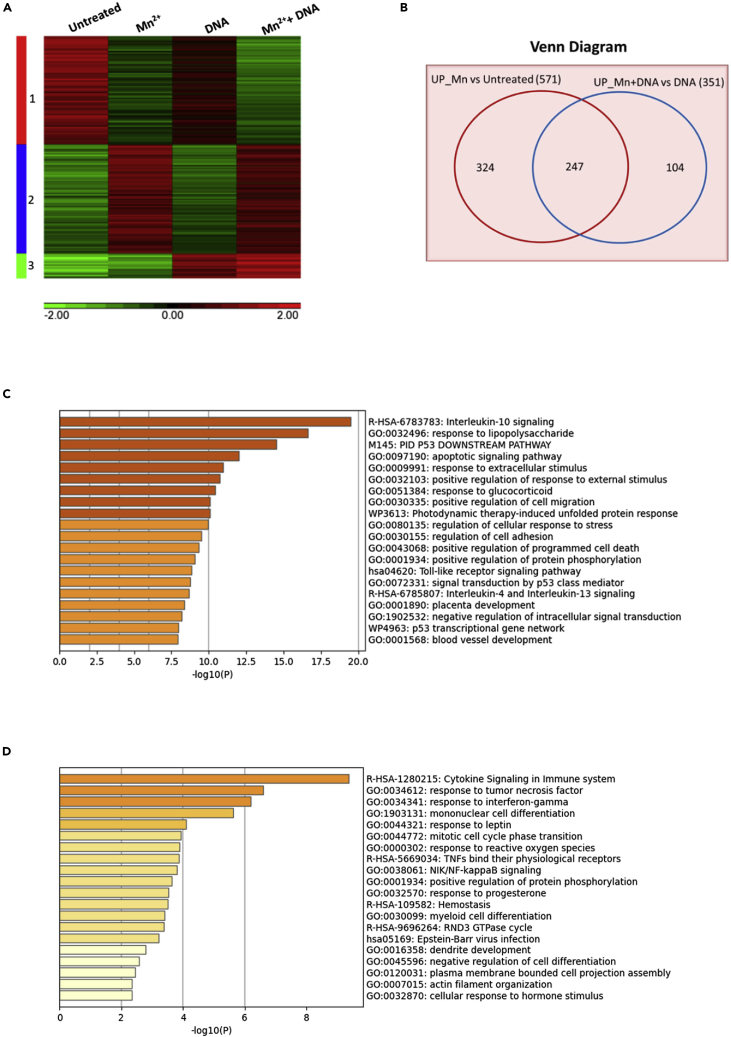
Figure 2.
Microarray analysis for the impact of Mn treatment
Human primary macrophages (n = 3) were treated as indicated: untreated, MnCl2 treatment alone, DNA transfection alone, or MnCl2 pretreatment with DNA transfection. The cells were collected for RNA extraction, and the RNA samples were used as the input for the microarray platform. The microarray data was analyzed using 2-way ANOVA. The genes were selected by p-value <=0.05 and absolute fold change >=2.
(A) Heat map showing gene expression profiles among different treatments: untreated, MnCl2-treated, DNA-transfected, and Mn- and DNA-treated cells.
(B) Venn diagram showing overlap of differentially upregulated genes in macrophages on different treatments. (571 genes from Mn vs. Untreated and 351 genes from Mn + DNA vs DNA treated)
(C) Functional enrichment analysis by Metascape for 247 Mn-mediated and upregulated genes.
(D) Functional enrichment analysis by Metascape for 104 DNA-mediated and Mn-upregulated genes.