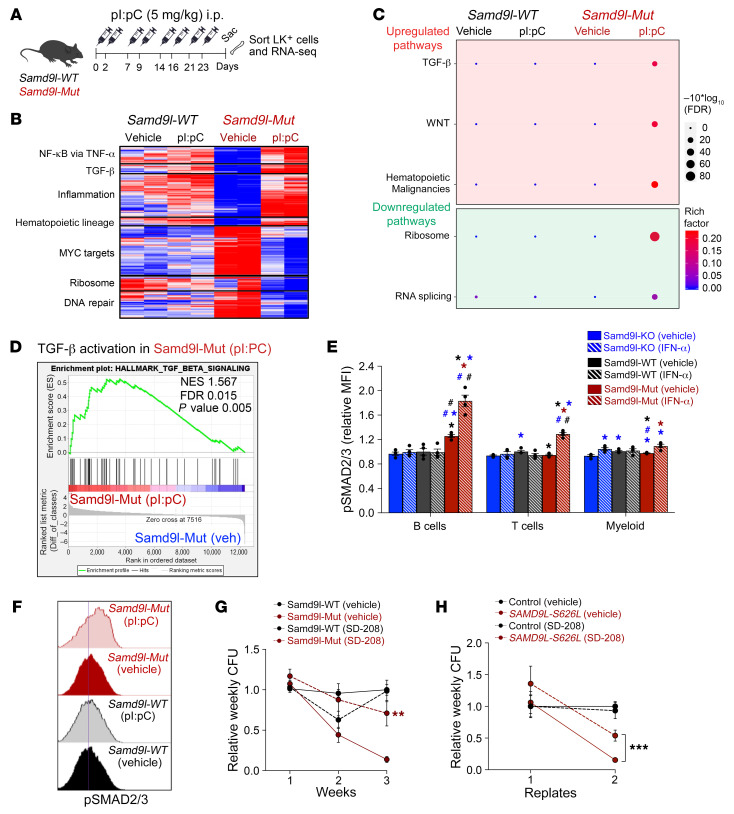
Figure 5. The lack of fitness in Samd9l-mutant cells is partly via TGF-β activation.
(A) Model of pI:pC treatment regimen followed by sorting the Lin–cKit+ (LK) population from Samd9l-WT and Samd9l-Mut mice to perform RNA-seq. (B) Heatmap showing the up- and downregulated pathways in Samd9l-WT and Samd9l-Mut treated with vehicle or pI:pC. (C) A plot of pathway enrichments of DEGs downregulated in pI:pC-treated Samd9l-Mut mice relative to vehicle-treated. The size of the circles represents gene counts, and the significance was determined by FDR. The color is dependent on the fold of enrichment. (D) Gene set enrichment analysis (GSEA) showing TGF-β pathway activation in pI:pC-treated Samd9l-Mut mice relative to vehicle-treated Samd9l-Mut mice. Normalized enrichment score (NES), FDR, and P value are indicated. (E) Phospho-SMAD2/3 expression in Samd9l-KO, Samd9l-WT, and Samd9l-Mut BM cells treated with IFN-α or vehicle (n = 4 per group). (F) Representative histograms of phospho-SMAD2/3 expression in B cells of Samd9l-WT and Samd9l-Mut cells after IFN-α or vehicle. (G and H) Serial CFU-C replating of Samd9l-WT and Samd9l-Mut cells (G) or human cells from a patient with SAMD9L-S626L mutation or control (H) with or without TGF-β inhibitor (SD-208). Data show at least 3 independent experiments. For panel E, Kruskal-Wallis test was used to perform an initial comparison across all groups and followed by pairwise comparisons with Wilcoxon’s rank-sum test. For panel G, for each genotype/time point, we used Wilcoxon’s rank-sum test to compare across the 2 treatments because data were not normally distributed. For panel H, a longitudinal mixed-effects regression model was used for statistical analysis followed by pairwise Tukey-adjusted tests evaluating the equality of means across each pair of groups at each time point. *P < 0.05, **P < 0.01, ***P < 0.001 compared with vehicle-treated groups. #P < 0.05 compared with pI:pC-treated groups. Error bars indicate the SEM for biological replicates. Blue, Samd9l-KO; black, Samd9l-WT; red, Samd9l-Mut (red); stripes, IFN-α or pI:pC; solid, vehicle. Color indicates the comparison group.