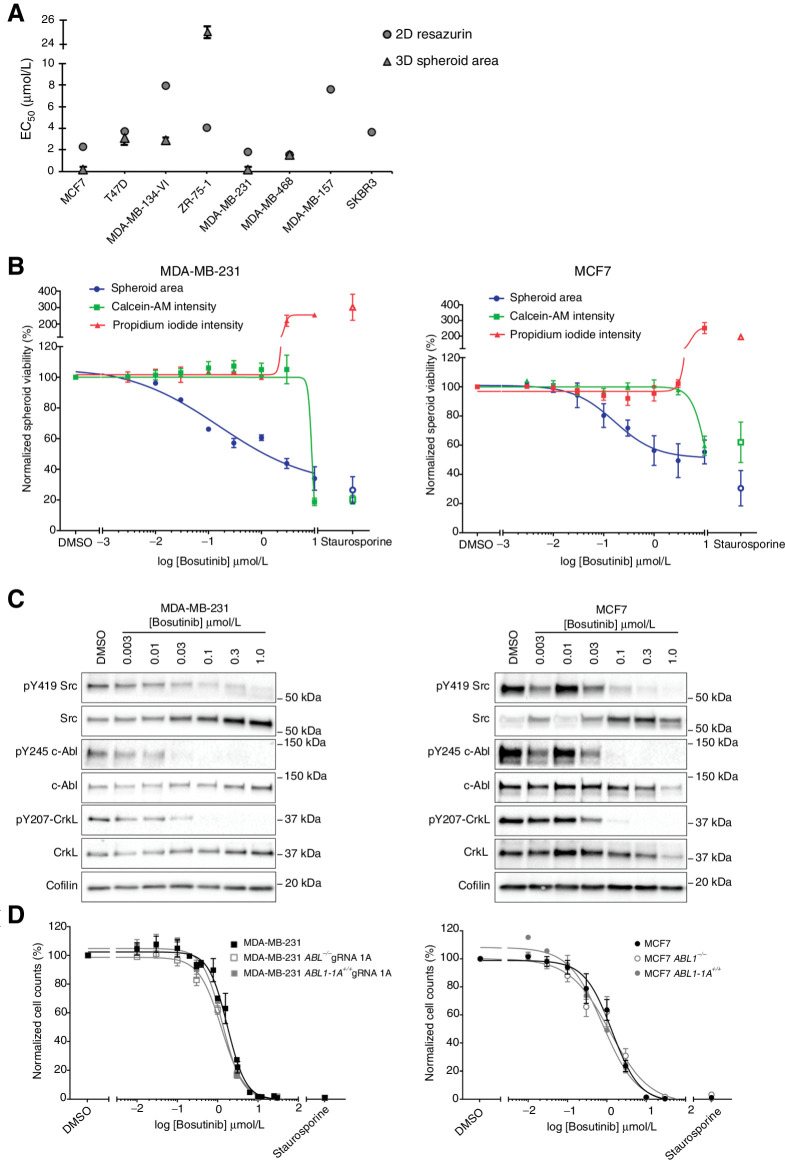
Figure 1.
Effect of bosutinib treatment on MDA-MB-231 and MCF7 cell lines. A, Bosutinib 2D and 3D EC50 values in a panel of breast cancer cell lines. MDA-MB-157 and SKBR3 are unable to form spheroids. B, 3D spheroid cell viability. Spheroid area, Calcein-AM intensity, and propidium iodide intensity were quantified using a custom CellProfiler pipeline. Staurosporine (1 μmol/L) was used as a positive control to induce cell death. C, Western blot analysis of cell lines grown as 3D spheroids. Data are representative of two independent experimental replicates. D, Loss of ABL1 does not alter bosutinib sensitivity. 2D bosutinib 8-point dilution for half-inhibitory concentrations (EC50). Additional 2D and 3D data are shown in Supplementary Fig. S4. All data shown are the mean of at least three independent experimental replicates except MDA-MB-134-VI and MDA-MB-468, which were two independent experimental replicates. ***, P < 0.0002; ****, P < 0.0001, as determined by a two-way ANOVA with Bonferroni multiple comparisons correction. All error bars are SEM.