Figure 4. In silico design of enzyme active sites.
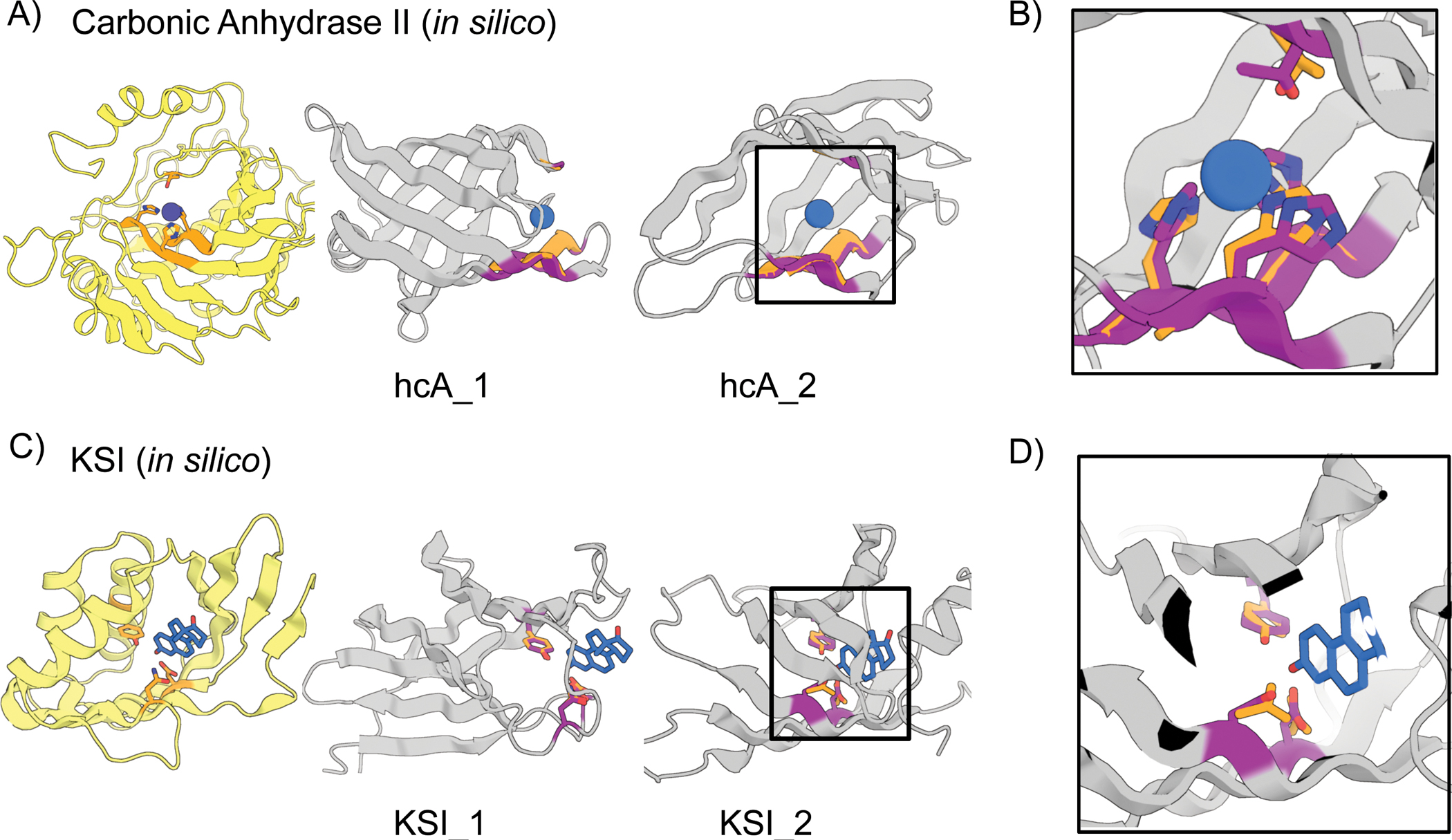
(A-B) Hallucinations using backbone description of site using RF. (C-D) Hallucination using sidechain description of site using AF2 augmented with trRosetta (Materials and Methods). (A) Carbonic anhydrase II active site (5YUI chain A residues 62–65, 93–97, 118–120). (B) Δ5-3-ketosteroid Isomerase active site (1QJG chain A residues 14, 38, 99). Colors: native protein scaffold (light yellow); native functional motif (orange); hallucinated scaffold (gray); hallucinated motif (purple); bound metal (blue). Active site residues shown for boxed designs in panel B and for carbonic anhydrase II, and Δ5-3-Ketosteroid Isomerase respectively. Design metrics (AF pLDDT, motif RMSD AF versus native): hcA_1 (73, 1.04 Å), hcA_2 (71, 0.62 Å), KSI_1 (84, 0.30 Å Cb), KSI_2 (72, 0.53 Å Cb)
