Figure 5. Design of protein-binding proteins.
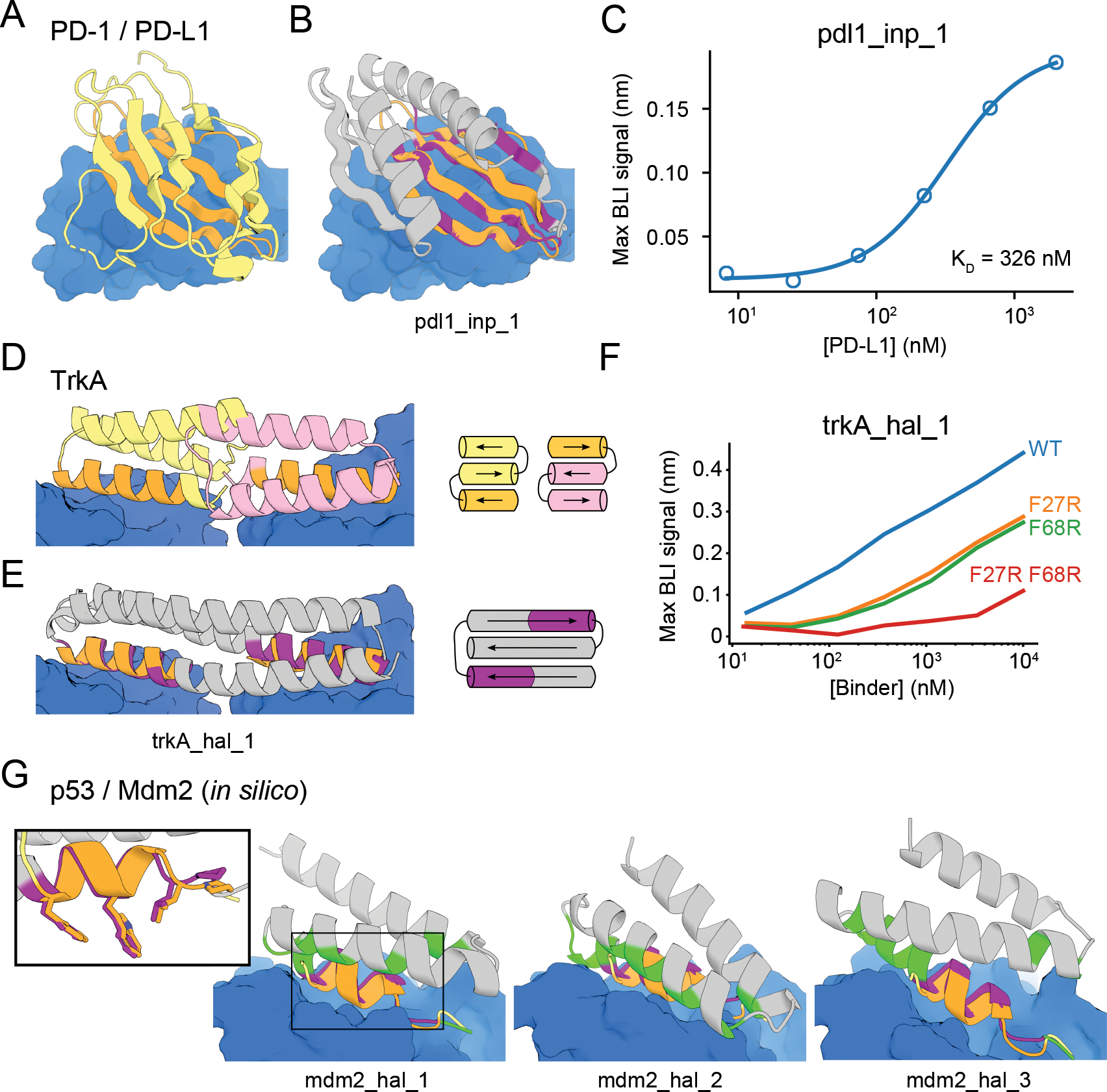
Designs containing target-binding interfaces built around native-complex-derived binding motifs. Targets are in blue, native scaffolds in yellow or pink, native motifs in orange, designed scaffolds in gray and designed motifs in purple. (A) Crystal structure of high-affinity consensus (HAC) PD-1 in complex with PD-L1. (B) Inpainted PD-L1 binder superimposed on PD-1 interface motif. (C) Max BLI binding signal versus PD-L1 concentration. (D) Crystal structure of previously designed TrkA minibinder in complex with TrkA, superimposed on TrkA receptor dimer. (E) Hallucinated bivalent TrkA binder. Protein topologies of (D-E) are shown to the right. (F) Max BLI binding signal versus TrkA concentration, showing that both binding sites bind TrkA. (G) Hallucinated Mdm2 binder designs superimposed on native p53 helix in complex with Mdm2 (see also Fig. S17D–E). New binding interactions (hallucinated residues within 5 Å of the target) are in green. Inset: Overlay of mdm2_hal_1 and native p53 helix showing key sidechains for binding.
