Figure 2.
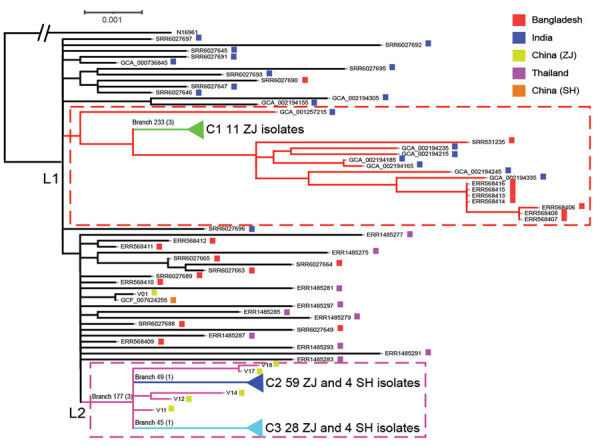
Maximum-likelihood phylogenetic tree of 161 Vibrio cholerae O139 (sequence type 69) isolates from Zhejiang Province, China, 1994–2018, and isolates from outside of China. The tree was rooted using the seventh pandemic O1 strain N16961 as an outgroup. Lineage 1 (L1) and lineage 2 (L2) are demarcated with red dashed lines and pink dashed-line boxes. The 3 clusters (C1, C2, and C3) are collapsed to reduce figure size (Appendix 2 Figure 2). Key branches are marked with a branch number followed in brackets by the number of single nucleotide polymorphisms that supported the branch. The colored solid squares at the end of isolate names indicate the location of isolation of the isolates. GenBank accession numbers were used as isolate names for O139 isolates not from Zhejiang Province. SH, Shanghai; ZJ, Zhejiang.
