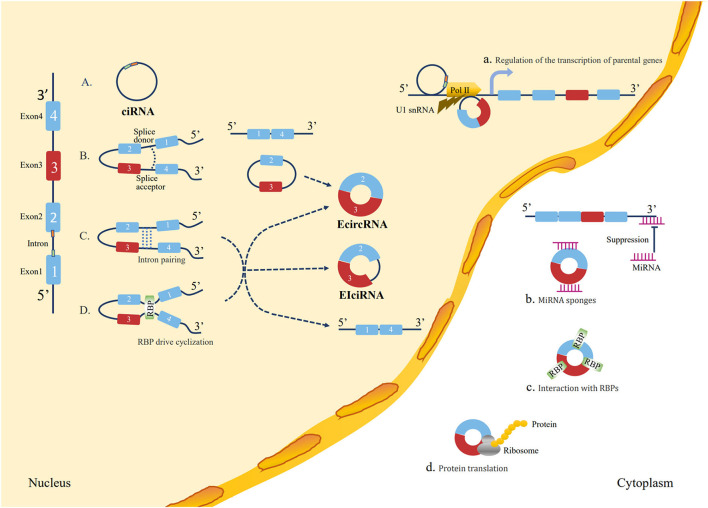
Figure 1.
Loop formation mechanisms and biological functions of circRNAs. Loop formation mechanisms: (A) ciRNA formation: the elements near the splice site escape debranching stably so that the intron lariat is formed from the splicing reaction. (B) Lariat-driven circularization: the 5′ splice donor of exon 1 and the 3′ splice acceptor of exon 4 link up end-to-end by exon skipping and form an exon-containing lariat structure. Finally, the ecircRNA forms after introns are removed. (C) Intron pairing-driven circularization: direct base pairing of introns forms a circulation structure, thereby forming ecircRNA or EIciRNA after intron removal. (D) RBP-driven cyclization: RBPs bridge two flanking introns close together and then remove introns to form circRNAs. Biological functions: (a) Regulation of the transcription of parental genes: circRNAs play a regulatory role in the transcription of their parent coding genes. (b) Function as miRNA “sponge”: circRNAs contain a common miRNA response element (MRE) that can bind to miRNA and prevent them from interacting with mRNA. (c) Interaction with RBPs: circRNAs can bind to RBPs to regulate mRNA expression by altering the splicing pattern or mRNA stability. (d) Protein translation: circRNAs have coding potential and can be translated into proteins with ribosomes.