Figure 2.
Multi-omic integrative analysis of the sex-dependent molecular responses to SARS-CoV-2 infection
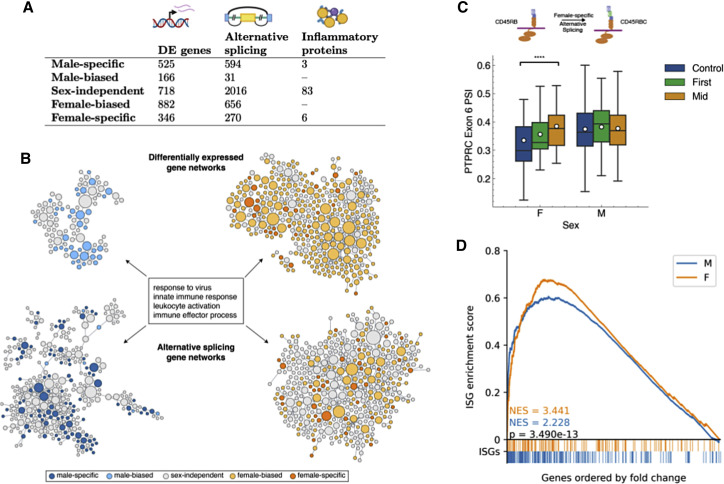
(A) The number of sex-dependent molecular variations upon SARS-CoV-2 infections discovered using gene expression, alternative splicing, and proteomics.
(B) Sex divergence of gene expression and alternative splicing visualized in the virus response module of a tissue-specific functional network. Each node represents a gene, with its size proportional to its connectedness in the network. Clusters shown here represent the immune response genes from males (left) and females (right) with differential expression (top) and differential alternative splicing events (bottom) during SARS-CoV-2 infection. This network is constructed by probabilistically integrating a compendium of thousands of public omics datasets to provide functional maps of biological processes and pathways. Projecting each sex-specific signature onto this network reveals significant enrichment of the immune response module in both gene expression and alternative splicing.
(C) Box-and-whisker plot of exon inclusion changes of PTPRC exon 6 upon SARS-CoV-2 infection in females (n = 55) and males (n = 255) throughout the infection course. The white circles represent the mean, the center line shows the median, and the upper and lower edges of the boxes represent the upper and lower quartiles, respectively. The differential splicing of PTPRC will alter the protein product of PTPRC, known as CD45, in females.
(D) ISGs are significantly enriched in both males and females during COVID-19 infection but significantly more so in females as shown by the higher normalized enrichment score (NES) from gene set enrichment analysis. Colored bars below the x axis represent ISG locations within the list of all differentially expressed genes, ordered by fold change from baseline samples. p value calculated using the Mann-Whitney U test.