Fig. 4.
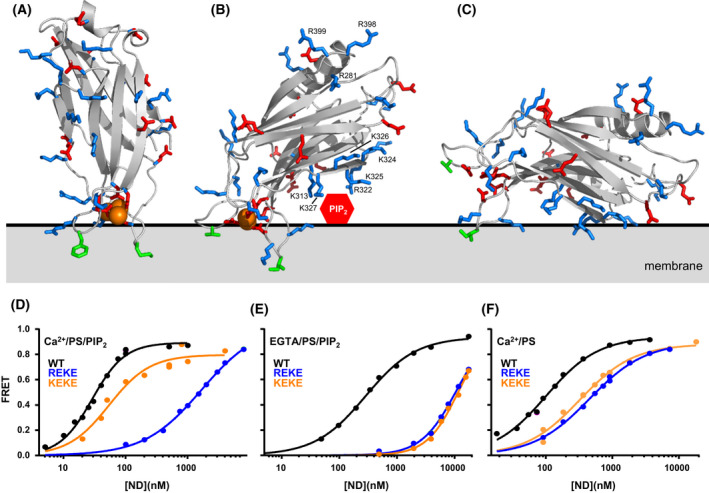
Structure and membrane interactions of the Synaptotagmin‐1 C2 domains. (A, B) Ribbon diagrams of the NMR structures of the Syt1 C2A (A) and C2B (B) domains [47, 51] bound to Ca2+ (orange spheres) (PDB accession numbers 1BYN and 1K5W, respectively) in the approximate orientations with respect to a flat phospholipid bilayer (gray) defined by EPR [105, 107]. The slightly slanted orientation of the C2B domain in (B) enables interactions of residues from the polybasic region with PIP2 head groups that protrude from the bilayer surface (red hexagon). Basic residues involved in SNARE complex and membrane interactions are labeled and shown by stick models. Hydrophobic side chains that insert into the membrane are shown as green stick models. Note that K313, R322, K325 and K327 in the polybasic face of the C2B domain can readily bind to PIP2 in these orientations whereas K324 and K326 are oriented away from the membrane, farther from the PIP2 head group. These observations explain the selective disruption of neurotransmitter release caused by mutations in the polybasic face [108, 112]. (C) Approximately parallel orientation of the C2B domain with respect to the membrane expected in the absence of Ca2+, which is supported by MD simulations [87]. (D–F) Selectively strong disruption of Syt1 C2AB binding to membranes by the R322E,K325E (REKE) mutation but not by the K324E,K326E (KEKE) mutation is observed in the presence of Ca2+ and PIP2 but not in the absence of Ca2+ and/or PIP2. The plots show binding of C2AB to nanodiscs containing PS and PIP2 (D, E) or PS but not PIP2 (F) in the presence of Ca2+ and 125 mm KCL (D, F) or EGTA and 50 mm KCl (E). These data were reported in [45]. The data in panel (E) were acquired at lower ionic strength because binding is very weak in the absence of Ca2+.
