Fig. 5.
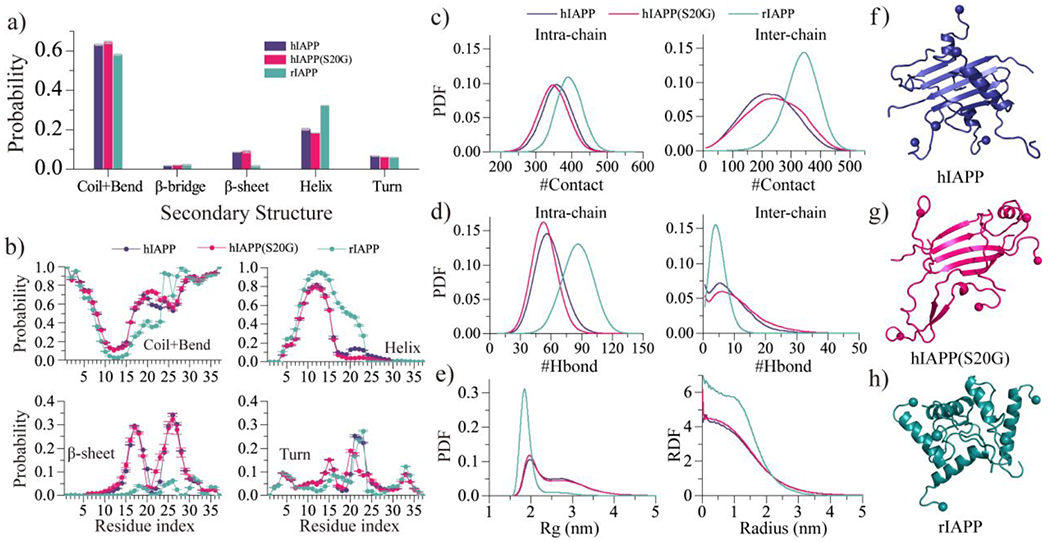
Oligomeric conformation analysis. a) The average secondary structure contents of unstructured (coil and bend), β-sheet, helix and turn conformation for the oligomers formed by hIAPP, hIAPP(S20G), and rIAPP peptides. b) The averaged propensity of every residue adopting coil and bend, helix, β-sheet, and bend conformations in five-peptide simulations for each type of amylin peptides. c-d) The probability distribution as a function of the number intrachain/interchain heavy-atom contact c) and backbone hydrogen bonds d). e) The probability distribution as a function of radius gyration (Rg) and radial distribution function (RDF) of Cα atoms for the self-assemblies formed by each peptide. f-h) The oligomeric formation of each type of amylin peptide. Only the last 500 ns trajectories from 60 independent simulations are used for the above conformational analysis.
