Graphical Abstract
Summary: Dairy calves are typically separated from the dam shortly after birth, but there is growing interest in rearing cows and calves together within dairy systems. Little work to date has evaluated differences in microbiota between conventionally reared and dam-reared dairy calves. We compared the fecal microbiota of 4-wk-old dairy calves reared conventionally and fed waste milk to that of calves reared with the dam. A principal coordinates analysis plot of β diversity, shown on the right, illustrates dissimilarity between the 2 treatment groups. Dam-reared calves showed higher relative abundance of Lactobacillus and lower relative abundance of taxa such as Bacteroides, Sutterella, and Prevotella. Calves permitted maternal contact also showed a predicted increase in l-tryptophan biosynthesis.
Highlights
-
•
We compared fecal microbiota in dam-reared and conventionally reared dairy calves, which were fed whole milk and waste milk, respectively.
-
•
Dairy calves reared with dam contact had higher relative abundance of Lactobacillus.
-
•
Conventionally reared calves had higher concentrations of taxa such as Bacteroides.
-
•
Dam-reared calves were predicted to have higher levels of l-tryptophan biosynthesis.
Abstract
The practice of rearing cows and calves together is gaining popularity on dairy farms, with different systems currently under assessment in mainland Europe, the United Kingdom, and Oceania. Research into the effects of cow–calf rearing has primarily focused on direct health and welfare implications, and little work has examined the role of different rearing paradigms on calf microbiota. We trialed a cow–calf rearing system on a Canadian dairy farm and compared fecal microbiota of these calves with the microbiota of calves reared according to the conventional practice of the same farm (separated from the dam and fed waste milk). At 4 wk, the conventionally reared calves had reduced relative abundance of Lactobacillus and higher relative abundance of other taxa, including Sutterella, Prevotella, and Bacteroides. We also detected predicted functional differences, such as reduced l-tryptophan biosynthesis in conventionally reared calves. These results suggest that maternal contact may influence the calf microbiota, but the observed differences are also likely related to other aspects of the rearing environment independent of maternal contact (e.g., potential exposure to antibiotic residues in waste milk). These findings provide preliminary evidence of the effects of early rearing environments on the establishment of the dairy calf fecal microbiota. This research is needed, given the critical role of the bovine gut microbiome in behavioral, metabolic, and immune development.
In mammalian offspring, the gut microbiota plays a critical role in modulating host enteric development, immune function, growth, and energy balance (Arrieta et al., 2014; Petersen et al., 2019). The effects of early rearing conditions on neonatal fecal microbiota have been investigated in several mammalian species, including humans (Gritz and Bhandari, 2015), rats (Macrì, 2016), and piglets (Maradiaga et al., 2018); comparatively little work has been done to understand the influence of rearing conditions on the calf gut microbiota (Malmuthuge and Guan, 2017). Neonatal dairy calves are particularly susceptible to enteric infection: scours, linked to dysbiosis in the gut microbiome, are a leading cause of dairy calf mortality (Uetake, 2013; Cho and Yoon, 2014). Further understanding of rearing conditions for dairy calves may aid in the establishment of protective microbial communities.
The dairy calf is the only mammalian farm animal to undergo systematic separation from the dam at birth. In conventional dairy operations, calves are typically separated within 12 h and often housed in individual pens or hutches in an effort to monitor colostrum intake and prevent exposure to pathogens (Vasseur et al., 2010; Windsor and Whittington, 2010). However, there is mounting interest in adopting dairy cow–calf rearing systems during the milk-feeding period (Johnsen et al., 2018). Early cow–calf separation could compound an already stressful period for the young calf, increasing the risk of microbial infection (Kouritzin and Guan, 2017). Because composition of the neonatal gut microbiome has significant influence on future health, brain development, and behavior (Diaz Heijtz, 2016), the implications of cow–calf separation on dairy calf microbiota warrants exploration.
In human infants, composition of the microbiome is heavily influenced by commensal bacterial transfer from the mother by means of skin contact and consumption of breast milk (Jost et al., 2014). Research on the establishment of fecal microbiota in lambs (Bi et al., 2019) has noted that feeding method (suckling the dam vs. bottle feeding) affects the sources of bacterial transmission from both the dam and the environment. In dairy calves, research has focused almost exclusively on conventionally reared (i.e., separated) animals, and bacterial species associated with weight gain, scours, and disease status have been identified (Oikonomou et al., 2013). Studies that have investigated specific early rearing conditions for dairy calves have mainly sought to examine differences in microbial populations as a result of differing diets and nutrition, (e.g., Dill-McFarland et al., 2019; Maynou et al., 2019). However, one recent study investigated differences in maturation of oral and fecal microbiota in maternally reared beef calves versus early-separated dairy calves, finding no demonstrable differences (Barden et al., 2020). Another study (Cunningham et al., 2018) identified differences in β diversity in the ruminal microbiome of dam-reared calves versus those fed milk replacer; further research is needed to determine the extent to which these differences are related to diet and whether this pattern is mirrored in the calf fecal microbiome.
The present study aimed to compare fecal microbial composition in dam-reared animals with that of artificially reared counterparts (separated and fed waste milk) on the same farm. We hypothesized that early rearing environment and the presence or absence of the dam would be associated with alterations in the fecal microbiota of preweaned dairy calves. The study (ethics approval no. A15-0082) was conducted at the University of British Columbia (UBC) Dairy Education and Research Centre (Agassiz, BC, Canada). Thirty-seven dairy calves were assigned prepartum to treatment group and enrolled at birth.
Calves in the maternal contact group (n = 22) remained with the dam in the maternity area for a minimum of 24 h after birth before transfer to the experimental setup. They were fed 4 L of quality-controlled colostrum within the first 6 h of life. Calves were then moved to a “calf creep,” a sawdust-bedded pen measuring approximately 10 × 3 m, where they remained during the day from 0630 to 1730 h. The sawdust was replaced as needed and completely refreshed on a weekly basis. Calves were offered unpasteurized whole milk twice per day (0900 and 1600 h) ad libitum. This milk was obtained in the milking parlor from a single trial cow to mimic milk that a nursing calf would obtain from the dam. (This cow was varied over the course of the trial due to the dynamic nature of cow–calf enrollment). The milk initially was offered via nipple bottles when the calves were <3 d old, and subsequently was offered from a milk bar. At approximately 1730 h each evening, calves were transferred to a freestall pen where their dams were housed. Calves were provided ad libitum access to the cow's hay and TMR during the night, and these rations were also provided in the calf creep and refreshed daily. Because this experiment was part of a larger study aimed at assessing dairy cow motivation (Wenker et al., 2020), the dams of 11 calves in this group wore udder nets to prevent suckling; however, calves could perform all other affiliative behaviors with their dams and could suckle from all other dams in the pen. Where possible, calves were randomly allocated to groups; however, some calves were born overnight, and the possibility that they had nursed could not be ruled out. Because these calves were not immediately separated, they were included in the cow–calf nursing group.
Calves in the conventional group (n = 15) were raised according to standard practice at the UBC Dairy Education and Research Centre (i.e., separated from the dam within 6 h and moved to individual cubicles in a calf barn located approximately 300 m away). Similar to calves in the maternal contact group, calves were fed 4 L of quality-controlled colostrum within the first 6 h of life. Subsequently, calves were fed pasteurized waste milk from the herd at an allowance of 4 L twice per day by bottle. On this farm, waste milk mostly consisted of transition cow milk (within 2–6 d after calving) supplemented by whole milk from the bulk tank. Milk from cows with high SCC was also included, as was milk from any cows treated with antimicrobials. At approximately 7 d of age, calves in this group were transferred to sawdust-bedded group pens (each 4.9 × 7.3 m) with up to 8 other calves. Sawdust was replaced as needed and completely refreshed on a weekly basis. In group pens, calves were fed pasteurized waste milk from automatic calf feeders at an allowance of 12 L/d and offered hay and grain (Hi-Pro Medicated, Hi-Pro Feeds) ad libitum.
Health events that occurred throughout the trial, including scours, elevated temperature, and coughing, were recorded for both groups of calves. Health was comparable across the groups, with 7 (32%) maternal contact calves and 7 (47%) conventional calves experiencing at least 1 adverse health event over the course of the trial. One maternal contact calf was treated with antibiotics (Resflor Gold, Intervet Inc.); however, exclusion of this calf from the analysis did not affect results.
Fecal samples were collected from calves when they were approximately 4 wk of age (mean ± SD = 26 ± 2.8 d). Following the afternoon feeding, samples were collected rectally using sterile palpation sleeves with lubricant (First Priority Inc.). The lubricant was subsequently used as a negative extraction control. Environmental samples were obtained at each visit by sampling bedding material, within the 2 d before bedding change, from different areas within the calf creep, freestall pen, and calf barn. All samples were collected in sterile palpation sleeves, transferred immediately into 2-mL microfuge tubes at the onsite laboratory, and frozen at −70°C.
At the end of the trial, samples were transported on dry ice to UBC's Michael Smith Laboratories for DNA extraction using a QIAamp PowerFecal DNA Kit (Qiagen Inc.) according to the manufacturer's instructions. Picogreen quantification ensured that samples had minimum DNA concentrations of 1 ng/µL. Samples were then shipped to Integrated Microbiome Resources (IMR; Dalhousie University, Halifax, NS, Canada) for sequencing. MiSeq Short Amplicon sequencing was performed to amplify the variable V4–V5 regions of the 16S sequencing target. The library was prepared according to the IMR protocol (see https://imr.bio/protocols.html). Demultiplexed sequencing data used in this study are deposited into the Sequence Read Archive of NCBI (accession no. PRJNA700665).
Data were analyzed using R (version 3.6.1; R Core Team) and Qiime 2 (Quantitative Insights into Microbial Ecology 2, version 2019.1; Bolyen et al., 2019). Predictive functional analysis was conducted using the PICRUSt2 tool (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; Douglas et al., 2020). Specifically, raw sequences were preprocessed in Qiime2 using a modified code from the IMR repository (Comeau et al., 2017). Exported files from Qiime2, and the metadata, were imported using Phyloseq in R.
Sample quality was high; the average raw sequencing depth was 32,822 reads per sample, or 21,984 reads (interquartile range: 15,268–27,229) after a quality control step (DADA2) was applied to remove erroneous reads. A total of 8,966 distinct bacterial and archaeal amplicon sequence variants (ASV) were identified in the 129 samples. The final feature count (after removing features appearing <0.1% of the mean sequencing depth across all samples) was 4,436 distinct ASV. As the lowest calf sample depth was 5,903 reads, samples were rarefied to a sequencing depth of 5,000 reads. There was no amplification in any negative controls.
For β diversity plots, samples were further log-transformed. Bray-Curtis principal coordinates analysis revealed dissimilarity in microbiota composition between maternal contact calves and conventional calves (P = 0.001, permutational multivariate ANOVA; Figure 1). Established communities of microbes are resilient to transformation (Dill-McFarland et al., 2019); thus, optimization of the early rearing environment could be critical to establishing the continued presence of beneficial microbes. The microbiota of conventionally reared calves in this study may be representative of calves reared under similar management practices in British Columbia and arguably elsewhere in the world.
Figure 1.

Principal coordinates analysis (PCoA) plot of Bray-Curtis dissimilarity in β diversity between treatment groups. Ellipses delineate 95% prediction areas for each group, with the dotted lines assuming a multivariate normal distribution and the solid lines assuming a multivariate t distribution. The red ellipse represents maternal contact calves, and the teal ellipse represents conventionally reared calves. The P-value corresponds to results from the adonis permutational multivariate ANOVA.
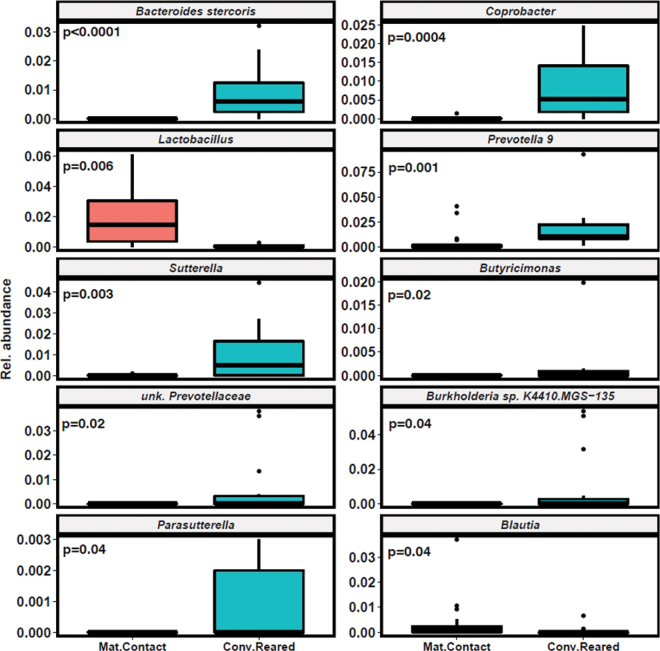
Raw abundances were transformed into relative abundances, and ASV were aggregated into species-level identification. We identified differences in relative abundance of specific bacterial taxa (Figure 2) using false discovery rate-corrected Mann-Whitney U tests. The conventionally reared calves had reduced relative abundance of Lactobacillus members (P = 0.003) and increases in other taxa such as Bacteroides stercoris (P < 0.001). Previous research has identified distinct taxonomic clusters in the preweaned calf ileum that are either Lactobacillus or Bacteroides dominant (Malmuthuge et al., 2019). Lactobacillus spp. have been shown to have probiotic effects for calves and offer protection against calf scours (Fernández et al., 2018). According to Malmuthuge et al. (2019), early exposure to Lactobacillus may prime the host (via initial expression of key proinflammatory chemokines) for future anti-inflammatory responses. Such early exposure could be helpful for neonatal calves due to naïve mucosal immune status. Maternal contact calves also had higher relative abundance of Blautia, which has been identified (e.g., in Jang et al., 2019) as a prominent genus in the gut of healthy calves.
Figure 2.
Relative abundance of significantly different bacterial taxa present within stool of maternal contact (Mat.Contact; n = 22) and conventionally reared (Conv.Reared; n = 15) calves. P-values represent results from false discovery rate-corrected Mann-Whitney U tests. Rel. = relative; unk. = unknown. The midlines represent median values for each group. The boxes represent interquartile range (IQR), whiskers are 1.5 × IQR, and dots represent any datapoints beyond 1.5 × IQR.
In addition to Bacteroides stercoris, conventionally reared calves showed increases in several other relevant taxa, including Coprobacter (P < 0.001), Prevotella 9 (P = 0.002), Sutterella (P = 0.003), Butyricimonas (P = 0.02), Parasutterella (P = 0.04), Burkholderia (P = 0.04), and unknown Prevotella (P = 0.04). Both Bacteroides and Burkholderia comprise significant pathogenic species with high antimicrobial resistance rates (Wexler, 2007; Rhodes and Schweizer, 2016); however, these taxa also predominate in healthy calves (Klein-Jöbstl et al., 2014; Borsanelli et al., 2018; Song et al., 2019). Similarly, Prevotella has been identified in the fecal microbiota of both diarrheic and healthy calves (Ma et al., 2020). Prevotella is a dominant genus in the ruminant gastrointestinal tract and is known to serve important functions in the rumen ecosystem, including production of short-chain fatty acids such as propionate and acetate (Dill-McFarland et al., 2017) and amino acids (Belanche et al., 2012). The abundance of Prevotella is influenced by the level of fiber in the diet (Klein-Jöbstl et al., 2014) and is thus particularly relevant during calf weaning, when milk is withdrawn and fiber-rich feeds such as hay and calf starter are introduced.
Further research aimed at identifying specific operational taxonomic units in calves reared with and without the dam could provide important insights into the observed difference in Prevotella. For example, one specific operational taxonomic unit has been found to be negatively correlated with ADG in calves (Dill-McFarland et al., 2017), and several Prevotella species are associated with periodontal disease in cattle (Borsanelli et al., 2018). Others, however, may simply be a function of the developmental stage of the rumen. Sutterella and Parasutterella are associated with gastrointestinal disease in humans, and Sutterella possess the ability to degrade IgA (Chen et al., 2018; Kaakoush, 2020). However, these effects have not been investigated in calves.
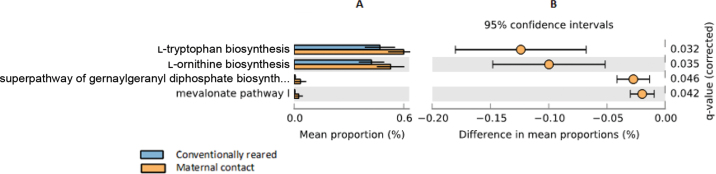
Difference in the taxonomic abundance between groups further resulted in changes within the predicted functional microbiomes (according to PICRUSt2 analysis; Figure 3). Conventionally reared calves were predicted to have reduced levels of l-tryptophan and l-ornithine biosynthesis (false discovery rate-corrected P-values of 0.032 and 0.035, respectively) in addition to reduced expression of the mevalonate pathway (P = 0.042) and the superpathway of geranylgeranyl diphosphate biosynthesis (P = 0.046). Tryptophan is a precursor of serotonin, which stimulates components of both the innate and adaptive immune systems and may enhance the calf's immune response to pathogens (Hernández-Castellano et al., 2018). Lactobacillus, which as previously described was more abundant in the calves allowed dam contact, has also been linked to greater tryptophan and l-ornithine metabolism within the gut (Qi et al., 2019). Collectively, these differences in both taxonomic composition and predicted functionality suggest potential advantages for calves reared with maternal contact compared with conventionally reared counterparts. However, the functional differences can only be predicted because the analyses are solely on 16S targets, and PiCRUST has been based on the human microbiome.
Figure 3.
Results of the PICRUSt2 (Douglas et al., 2020) predicted functional analysis. Blue bars represent samples from conventionally reared calves, and orange bars represent samples from maternal contact calves. Relevant metabolic pathways are shown on the left y-axis. Panel A displays mean proportion (%) on the x-axis, and panel B displays the difference in mean proportions (%) on the x-axis. Error bars represent 95% CI. False discovery rate-corrected P-values (q-values) are shown on the right y-axis.
Additionally, we cannot be certain to what extent maternal contact itself influenced changes in relative abundance of taxa and associated functional differences. There were no observable differences in α diversity (P > 0.05 in Wilcoxon rank sum tests, according to the Shannon diversity index). With regard to differences in β diversity, it is likely that these factors were also affected by environment and diet. To investigate the effect of environment, we compared the microbiota in environmental samples using Bray-Curtis principal coordinates analysis. The microbiota composition for the calf barn, in which the conventional calves were housed, differed from that of the area in which the maternal contact calves were housed (calf creep during the day and freestall pen at night; P = 0.01 permutational multivariate ANOVA). Comparison between the calf creep and the calf barn yielded a nonsignificant tendency (P = 0.07). Taxa-specific differences in environmental samples also showed tendencies (P ≤ 0.1 > 0.05) for higher concentrations of Coprobacter and Prevotella 9 in the calf barn compared with the calf creep and for lower concentrations of Lactobacillus. Thus, differences in the environment appeared to reinforce the differences between the groups to some extent. The inverse, however, is equally plausible: that the calves themselves led to environmental differences, given that the groups had different microbiota.
Several other important questions remain to be answered. For instance, the maternal contact calves in our study were group housed during the day, with collective nighttime access to their dams. As this housing likely involved regular reciprocal licking and other affiliative behaviors between dams and calves, we may have expected a high degree of homogeneity of microbiota between the calves, as social transmission of microbiota has been observed in other species (Münger et al., 2018). The data presented in Figure 1, however, do not seem to support this conclusion. Previous research in which calves were housed in a manner similar to that in the current study (dynamically, with nighttime access to their dams) suggests that calves spend more time nursing their own dam compared with other dams (Johnsen et al., 2015) and that, regardless of suckling opportunity, dam–calf pairs engage in more affiliative behaviors with each other relative to other animals in the same group (Johnsen et al., 2015).
To tease apart the nuances of social microbiota transmission from dam to calf, future studies could house calves individually with their respective dams. This experimental design may have limited feasibility in commercial contexts, and it is unknown, from a welfare perspective, whether dam–calf contact alone is an adequate substitute for the welfare benefits afforded by interactions between calves (Costa et al., 2016). One advantage of the current study design is that it more closely mimics the natural herd dynamics exhibited by semi-wild cattle (Vitale et al., 1986) and may be a more feasible option for implementation on commercial dairy farms.
Research on vertical transfer of microbes from ruminant milk is limited (Oikonomou et al., 2020). In humans, the direct contribution of milk microbiota to the infant has been estimated at a modest 4.9%, with additional indirect contributions hypothesized (Williams et al., 2019). The conventionally reared calves in our study were fed waste milk from the herd (albeit pasteurized), which may have contained antibiotic residues; this diet could have contributed to increased prevalence of genera associated with antimicrobial resistance. The literature provides conflicting messages on the effects of feeding calves milk containing antimicrobials. For example, Feng et al. (2020) found no difference in fecal microbiota of calves fed whole milk with and without pirlimycin when compared across the Comprehensive Antibiotic Resistance Database. Conversely, Van Vleck Pereira et al. (2014) found that fecal samples from calves fed raw milk containing drug residues (e.g., from ceftiofur, ampicillin, penicillin, and oxytetracycline) had a higher proportion of multidrug-resistant Escherichia coli. In a follow-up study (Van Vleck Pereira et al., 2016), the microbiota of calves fed raw milk with and without drug residues could be distinguished at the genus level. Waste milk feeding is common practice on North American dairy farms, with the percentage of herds feeding waste milk ranging from approximately 48 to 78% depending on region (Vasseur et al., 2010; USDA, 2016). It should be noted that feeding calves milk from treated cows does not represent best practice (Aust et al., 2013), but as long as this practice remains common, further research is needed to understand its long-term implications on the calf microbiome.
In their review, Malmuthuge and Guan (2017) emphasized the importance of understanding maternal factors and their effects on the dairy calf microbiome. These authors were alluding to maternal factors such as stress during gestation, methods of delivery, and maternal genetics. However, the presence of the dam herself as a maternal factor merits consideration, particularly in light of the increasing popularity of dairy cow–calf rearing systems and associated effects on welfare and health (Johnsen et al., 2018; Beaver et al., 2019; Meagher et al., 2019). Future research should aim to better to separate the influence of maternal contact from diet and environment. This effort could be facilitated by increasing the size and number of relevant groups to create a factorial design. There is a pseudoreplication issue inherent in housing different treatment groups in separate environments; however, there is also evidence of horizontal transmission of microbiota in mammalian species in shared environments, including baboons (Tung et al., 2015), chimpanzees (Moeller et al., 2016), and humans and canines (Song et al., 2013). Although transmission modes of gut microbiota in mammals are still poorly understood, it is likely that a shared environment produces homogenizing effects. A delicate balance is therefore needed in future studies to minimize the effects of pseudoreplication while simultaneously mitigating the homogenizing effect of housing multiple groups of calves in the same environment.
Recent work with a small number of beef and dairy calves (Barden et al., 2020) has suggested a greater phylogenetic similarity between calf fecal microbiota and the oral (vs. fecal and milk) microbiota of cows. In contrast, the early gut microbes of lambs permitted to nurse were derived primarily from the teat skin of their dams (Bi et al., 2019). If the presence of the dam is indeed found to be a significant factor, maternal contact itself could be further deconstructed to establish the primary sources of calves' microbiota.
Notes
The animal work was funded by Natural Sciences and Engineering Research Council of Canada (NSERC; Ottawa, ON, Canada) Discovery grants to DW and MvK. The bacteriological analysis was funded by The Hans Sigrist Research Prize, awarded to MvK.
We thank the students at the University of British Columbia (UBC) Dairy Education and Research Centre (Agassiz, BC, Canada) for their care of the study's calves and Wali Sahar (UBC) for his assistance with sample collection.
Author contributions: conceptualization, AB, DW, and MvK; field work: AB; DNA extraction: AB and CP; analysis: CP; draft preparation, AB; draft editing, AB, CP, DW, BF, and MvK.
The authors declare no known conflicts of interests.
References
- Arrieta M.C., Stiemsma L.T., Amenyogbe N., Brown E., Finlay B. The intestinal microbiome in early life: Health and disease. Front. Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. 25250028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aust V., Knappstein K., Kunz H.J., Kaspar H., Wallmann J., Kaske M. Feeding untreated and pasteurized waste milk and bulk milk to calves: Effects on calf performance, health status and antibiotic resistance of faecal bacteria. J. Anim. Physiol. Anim. Nutr. (Berl.) 2013;97:1091–1103. doi: 10.1111/jpn.12019. 23205592. [DOI] [PubMed] [Google Scholar]
- Barden M., Richards-Rios P., Ganda E., Lenzi L., Eccles R., Neary J., Oultram J., Oikonomou G. Maternal influences on oral and faecal microbiota maturation in neonatal calves in beef and dairy production systems. Anim. Microbiome. 2020;2:31. doi: 10.1186/s42523-020-00049-1. 33499967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver A., Meagher R.K., von Keyserlingk M.A.G., Weary D.M. Invited review: A systematic review of the effects of early separation on dairy cow and calf health. J. Dairy Sci. 2019;102:5784–5810. doi: 10.3168/jds.2018-15603. 31079908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanche A., Doreau M., Edwards J.E., Moorby J.M., Pinloche E., Newbold C.J. Shifts in the rumen microbiota due to the type of carbohydrate and level of protein ingested by dairy cattle are associated with changes in rumen fermentation. J. Nutr. 2012;142:1684–1692. doi: 10.3945/jn.112.159574. 22833657. [DOI] [PubMed] [Google Scholar]
- Bi Y., Cox M.S., Zhang F., Suen G., Zhang N., Tu Y., Diao Q. Feeding modes shape the acquisition and structure of the initial gut microbiota in newborn lambs. Environ. Microbiol. 2019;21:2333–2346. doi: 10.1111/1462-2920.14614. 30938032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsanelli A.C., Lappin D.F., Viora L., Bennett D., Dutra I.S., Brandt B.W., Riggio M.P. Microbiomes associated with bovine periodontitis and oral health. Vet. Microbiol. 2018;218:1–6. doi: 10.1016/j.vetmic.2018.03.016. 29685214. [DOI] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. 31341288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-J., Wu H., Wu S.-D., Lu N., Wang Y.-T., Liu H.-N., Dong L., Liu T.-T., Shen X.-Z. Parasutterella, in association with irritable bowel syndrome and intestinal chronic inflammation. J. Gastroenterol. Hepatol. 2018;33:1844–1852. doi: 10.1111/jgh.14281. 29744928. [DOI] [PubMed] [Google Scholar]
- Cho Y.-I., Yoon K.-J. An overview of calf diarrhea—Infectious etiology, diagnosis, and intervention. J. Vet. Sci. 2014;15:1–17. doi: 10.4142/jvs.2014.15.1.1. 24378583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau A.M., Douglas G.M., Langille M. Microbiome Helper: A custom and streamlined workflow for microbiome research. mSystems. 2017;2:e00127–e00136. doi: 10.1128/mSystems.00127-16. 28066818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J.H.C., von Keyserlingk M.A.G., Weary D.M. Invited review: Effects of group housing of dairy calves on behavior, cognition, performance, and health. J. Dairy Sci. 2016;99:2453–2467. doi: 10.3168/jds.2015-10144. 26874423. [DOI] [PubMed] [Google Scholar]
- Cunningham H.C., Austin K.J., Cammack K.M. Influence of maternal factors on the rumen microbiome and subsequent host performance. Transl. Anim. Sci. 2018;2(Suppl. 1):S101–S105. doi: 10.1093/tas/txy058. 32704752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R. Fetal, neonatal, and infant microbiome: Perturbations and subsequent effects on brain development and behavior. Semin. Fetal Neonatal Med. 2016;21:410–417. doi: 10.1016/j.siny.2016.04.012. 27255860. [DOI] [PubMed] [Google Scholar]
- Dill-McFarland K.A., Breaker J.D., Suen G. Microbial succession in the gastrointestinal tract of dairy cows from 2 weeks to first lactation. Sci. Rep. 2017;7 doi: 10.1038/srep40864. 28098248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill-McFarland K.A., Weimer P.J., Breaker J.D., Suen G. Diet influences early microbiota development in dairy calves without long-term impacts on milk production. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/AEM.02141-18. 30367001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. 32483366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Littier H.M., Knowlton K.F., Garner E., Pruden A. The impacts of feeding milk with antibiotics on the fecal microbiome and antibiotic resistance genes in dairy calves. Can. J. Anim. Sci. 2020;100:69–76. doi: 10.1139/cjas-2018-0202. [DOI] [Google Scholar]
- Fernández S., Fraga M., Silveyra E., Trombert A.N., Rabaza A., Pla M., Zunino P. Probiotic properties of native Lactobacillus spp. strains for dairy calves. Benef. Microbes. 2018;9:613–624. doi: 10.3920/BM2017.0131. 29633640. [DOI] [PubMed] [Google Scholar]
- Gritz E.C., Bhandari V. The human neonatal gut microbiome: A brief review. Front Pediatr. 2015;3:17. doi: 10.3389/fped.2015.00017. 25798435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Castellano L.E., Özçelik R., Hernandez L.L., Bruckmaier R.M. Short communication: Supplementation of colostrum and milk with 5-hydroxy-l-tryptophan affects immune factors but not growth performance in newborn calves. J. Dairy Sci. 2018;101:794–800. doi: 10.3168/jds.2017-13501. 29102139. [DOI] [PubMed] [Google Scholar]
- Jang J.Y., Kim S., Kwon M.S., Lee J., Yu D.H., Song R.H., Choi H.J., Park J. Rotavirus-mediated alteration of gut microbiota and its correlation with physiological characteristics in neonatal calves. J. Microbiol. 2019;57:113–121. doi: 10.1007/s12275-019-8549-1. 30456757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen J.F., de Passille A.M., Mejdell C.M., Bøe K.E., Grøndahl A.M., Beaver A., Rushen J., Weary D.M. The effect of nursing on the cow–calf bond. Appl. Anim. Behav. Sci. 2015;163:50–57. doi: 10.1016/j.applanim.2014.12.003. [DOI] [Google Scholar]
- Johnsen J.F., Mejdell C.M., Beaver A., de Passillé A.M., Rushen J., Weary D.M. Behavioural responses to cow-calf separation: The effect of nutritional dependence. Appl. Anim. Behav. Sci. 2018;201:1–6. doi: 10.1016/j.applanim.2017.12.009. [DOI] [Google Scholar]
- Jost T., Lacroix C., Braegger C.P., Rochat F., Chassard C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014;16:2891–2904. doi: 10.1111/1462-2920.12238. 24033881. [DOI] [PubMed] [Google Scholar]
- Kaakoush N.O. Sutterella species, IgA-degrading bacteria in ulcerative colitis. Trends Microbiol. 2020;28:519–522. doi: 10.1016/j.tim.2020.02.018. 32544438. [DOI] [PubMed] [Google Scholar]
- Klein-Jöbstl D., Schornsteiner E., Mann E., Wagner M., Drillich M., Schmitz-Esser S. Pyrosequencing reveals diverse fecal microbiota in Simmental calves during early development. Front. Microbiol. 2014;5:622. doi: 10.3389/fmicb.2014.00622. 25452753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouritzin V.A., Guan L. The colonization and establishment of the neonatal mammalian microbiome. Fine Focus. 2017;3:89–99. doi: 10.33043/FF.3.2.89-99. [DOI] [Google Scholar]
- Ma T., Villot C., Renaud D., Skidmore A., Chevaux E., Steele M., Guan L.L. Linking perturbations to temporal changes in diversity, stability, and compositions of neonatal calf gut microbiota: Prediction of diarrhea. ISME J. 2020;14:2223–2235. doi: 10.1038/s41396-020-0678-3. 32444812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrì S. Neonatal corticosterone administration in rodents as a tool to investigate the maternal programming of emotional and immune domains. Neurobiol. Stress. 2016;6:22–30. doi: 10.1016/j.ynstr.2016.12.001. 28229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmuthuge N., Guan L.L. Understanding the gut microbiome of dairy calves: Opportunities to improve early-life gut health. J. Dairy Sci. 2017;100:5996–6005. doi: 10.3168/jds.2016-12239. 28501408. [DOI] [PubMed] [Google Scholar]
- Malmuthuge N., Liang G., Griebel P.J., Guan L.L. Taxonomic and functional compositions of the small intestinal microbiome in neonatal calves provide a framework for understanding early life gut health. Appl. Environ. Microbiol. 2019;85:e02534–18. doi: 10.1128/AEM.02534-18. 30658973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maradiaga N., Aldridge B., Zeineldin M., Lowe J. Gastrointestinal microbiota and mucosal immune gene expression in neonatal pigs reared in a cross-fostering model. Microb. Pathog. 2018;121:27–39. doi: 10.1016/j.micpath.2018.05.007. 29742464. [DOI] [PubMed] [Google Scholar]
- Maynou G., Chester-Jones H., Bach A., Terré M. Feeding pasteurized waste milk to preweaned dairy calves changes fecal and upper respiratory tract microbiota. Front. Vet. Sci. 2019;6:159. doi: 10.3389/fvets.2019.00159. 31245388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R.K., Beaver A., Weary D.M., von Keyserlingk M.A.G. Invited review: A systematic review of the effects of prolonged cow–calf contact on behavior, welfare, and productivity. J. Dairy Sci. 2019;102:5765–5783. doi: 10.3168/jds.2018-16021. 31103300. [DOI] [PubMed] [Google Scholar]
- Moeller A.H., Foerster S., Wilson M.L., Pusey A.E., Hahn B.H., Ochman H. Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv. 2016;2 doi: 10.1126/sciadv.1500997. 26824072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münger E., Montiel-Castro A.J., Langhans W., Pacheco-López G. Reciprocal interactions between gut microbiota and host social behavior. Front. Integr. Neurosci. 2018;12:21. doi: 10.3389/fnint.2018.00021. 29946243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G., Addis M.F., Chassard C., Nader-Macias M.E.F., Grant I., Delbès C., Bogni C.I., Le Loir Y., Even S. Milk microbiota: What are we exactly talking about? Front. Microbiol. 2020;11:60. doi: 10.3389/fmicb.2020.00060. 32117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G., Teixeira A.G.V., Foditsch C., Bicalho M.L., Machado V.S., Bicalho R.C. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063157. 23646192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C., Bell R., Klag K.A., Lee S.H., Soto R., Ghazaryan A., Buhrke K., Atakan Ekiz H., Ost K.S., Boudina S., O'Connell R.M., Cox J.E., Villanueva C.J., Zac Stephens W., Round J.L. T cell–mediated regulation of the microbiota protects against obesity. Science. 2019;365 doi: 10.1126/science.aat9351. 31346040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Li Y., Yun H., Zhang T., Huang Y., Zhou J., Yan H., Wei J., Liu Y., Zhang Z., Gao Y., Che Y., Su X., Zhu D., Zhang Y., Zhong J., Yang R. Lactobacillus maintains healthy gut mucosa by producing l-ornithine. Commun. Biol. 2019;2:171. doi: 10.1038/s42003-019-0424-4. 31098404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes K.A., Schweizer H.P. Antibiotic resistance in Burkholderia species. Drug Resist. Updat. 2016;28:82–90. doi: 10.1016/j.drup.2016.07.003. 27620956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.J., Lauber C., Costello E.K., Lozupone C.A., Humphrey G., Berg-Lyons D., Gregory Caporaso J., Knights D., Clemente J.C., Nakielny S., Gordon J.I., Fierer N., Knight R. Cohabiting family members share microbiota with one another and with their dogs. eLife. 2013;2 doi: 10.7554/eLife.00458. 23599893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Malmuthuge N., Li F., Guan L.L. Colostrum feeding shapes the hindgut microbiota of dairy calves during the first 12 h of life. FEMS Microbiol. Ecol. 2019;95:203. doi: 10.1093/femsec/fiy203. 30307547. [DOI] [PubMed] [Google Scholar]
- Tung J., Barreiro L.B., Burns M.B., Grenier J.C., Lynch J., Grieneisen L.E., Altmann J., Alberts S.C., Blekhman R., Archie E.A. Social networks predict gut microbiome composition in wild baboons. eLife. 2015;4 doi: 10.7554/eLife.05224. 25774601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake K. Newborn calf welfare: A review focusing on mortality rates. Anim. Sci. J. 2013;84:101–105. doi: 10.1111/asj.12019. 23384350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA Dairy Cattle Management Practices in the United States, 2014. 2016. https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy14/Dairy14_dr_PartIII.pdf
- Van Vleck Pereira R., Lima S., Siler J.D., Foditsch C., Warnick L.D., Bicalho R.C. Ingestion of milk containing very low concentration of antimicrobials: Longitudinal effect on fecal microbiota composition in preweaned calves. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147525. 26808865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vleck Pereira R.V., Siler J.D., Bicalho R.C., Warnick L.D. In vivo selection of resistant E. coli after ingestion of milk with added drug residues. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115223. 25506918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur E., Borderas F., Cue R.I., Lefebvre D., Pellerin D., Rushen J., Wade K.M., de Passillé A.M. A survey of dairy calf management practices in Canada that affect animal welfare. J. Dairy Sci. 2010;93:1307–1315. doi: 10.3168/jds.2009-2429. 20172250. [DOI] [PubMed] [Google Scholar]
- Vitale A.F., Tenucci M., Papini M., Lovari S. Social behaviour of the calves of semi-wild Maremma cattle, Bos primigenius taurus. Appl. Anim. Behav. Sci. 1986;16:217–231. doi: 10.1016/0168-1591(86)90115-2. [DOI] [Google Scholar]
- Wenker M.L., Bokkers E.A.M., Lecorps B., von Keyserlingk M.A.G., van Reenen C.G., Verwer C.M., Weary D.M. Effect of cow–calf contact on cow motivation to reunite with their calf. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-70927-w. 32859980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. 17934076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.E., Carrothers J.M., Lackey K.A., Beatty N.F., Brooker S.L., Peterson H.K., Steinkamp K.M., York M.A., Shafii B., Price W.J., McGuire M.A., McGuire M.K. Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J. Nutr. 2019;149:902–914. doi: 10.1093/jn/nxy299. 31063198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor P.A., Whittington R.J. Evidence for age susceptibility of cattle to Johne's disease. Vet. J. 2010;184:37–44. doi: 10.1016/j.tvjl.2009.01.007. 19246220. [DOI] [PubMed] [Google Scholar]