Abstract
Background and Aims
Microbiome dysbiosis is associated with inflammatory destruction in Crohn’s disease [CD]. Although gut microbiome dysbiosis is well established in CD, the oral microbiome is comparatively under-studied. This study aims to characterize the oral microbiome of CD patients with/without oral manifestations.
Methods
Patients with CD were recruited with age-, gender- and race-matched controls. Potential confounders such as dental caries and periodontal condition were recorded. The oral microbiome was collected using saliva samples. Microbial DNA was extracted and sequenced using shotgun sequencing. Metagenomic taxonomic and functional profiles were generated and analysed.
Results
The study recruited 41 patients with CD and 24 healthy controls. Within the CD subjects, 39.0% had oral manifestations with the majority presenting with cobblestoning and/or oral ulcers. Principal coordinate analysis demonstrated distinct oral microbiome profiles between subjects with and without CD, with four key variables responsible for overall oral microbiome variance: [1] diagnosis of CD, [2] concomitant use of steroids, [3] concomitant use of azathioprine and 4] presence of oral ulcers. Thirty-two significant differentially abundant microbial species were identified, with the majority associated with the diagnosis of CD. A predictive model based on differences in the oral microbiome found that the oral microbiome has strong discriminatory function to distinguish subjects with and without CD [AUROC 0.84]. Functional analysis found that an increased representation of microbial enzymes [n = 5] in the butyrate pathway was positively associated with the presence of oral ulcers.
Conclusions
The oral microbiome can aid in the diagnosis of CD and its composition was associated with oral manifestations.
Keywords: Crohn disease, oral manifestations, oral microbiome
1. Introduction
Crohn’s disease [CD] is an inflammatory bowel disease [IBD] characterized by chronic inflammation of any part of the gastrointestinal tract. The cause of CD is unclear, with the key factors implicated in its pathogenesis including host genetics, immune dysregulation and gut microbiota alterations.1 CD has been strongly associated with an overall drop in microbiota species richness,2–4 whereby paediatric CD patients were found to have increased abundances of Enterobacteriaceae, Pasteurellacaea, Veillonellaceae and Fusobacteriaceae.5
Recently, metagenomic and metatranscriptomic studies on the gut microbiome have provided detailed information on this dysbiosis and proposed new treatment modalities.6 Perhaps unexpectedly, an abundance of oral microbes was found in the gut microbiome of IBD patients.7 Together with ectopic colonization of oral microbes, a recent study finding oral microbes of similar strains in the gut and oral microbiome of active CD patients,8 this suggests a pathogenic role for oral microbes. It has thus been postulated that in IBD, oral-disease bacteria expand in the IBD intestines through a sequential multi-stage approach9: [1] enhanced abundance and virulence of oral disease-associated bacteria with a corresponding reduction in intestinal colonization resistance, [2] translocation of oral bacteria to the intestines, and [3] colonization of the intestines by these oral disease-associated bacteria and exacerbation of disease in IBD. The notion that IBD can be driven by oral bacteria is consistent with reports that IBD patients have increased proportions of active periodontal disease,10 as well as common inflammasome-mediated inflammation pathways implicated in both periodontitis and IBD.11
While CD can affect any segment of the gastrointestinal tract, from the anus to the lips,12 extraintestinal manifestations can occur and frequently involve joints, skin, eyes and the oral mucosa. Oral manifestations such as oral aphthous ulcers are frequently associated with active disease and improve with the resolution of intestinal inflammation.13 Although oral manifestations can occur in both forms of IBD, it is more common in CD, with almost 50% of subjects reporting oral manifestations.14 Oral manifestations in CD can present as mucogingivitis, mucosal tags, deep ulceration, cobblestoning and lip swelling. They tend to be under-reported and typically to occur more often in paediatric patients.15 Sometimes, it can even be the first presenting sign in new cases of CD.16 Despite the apparent association between oral-associated microbiota and IBD, data regarding oral-associated microbiota in CD is limited. Furthermore, previous studies profiling the oral microbiome in CD cohorts did not account for oral manifestations, such as aphthous ulcers,13 mucogingivitis, mucosal tags, deep ulceration, cobblestoning and lip swelling, all of which are common in CD,14,15 and may have confounding effects on the composition of the oral microbiome.
Further obfuscating the role of the oral microbiome in CD is the issue of different sampling sites in the literature, such as the tongue,17 plaque,18 biopsies19 and saliva.20–22 Of the limited literature on the oral microbiome in CD, the saliva microbiome was most commonly examined and likely to have the greatest impact on the progression of CD.7 In those studies, Said et al. found an increased prevalence of Prevotella and Veillonella, but decreased Streptococcus and Haemophilus, in CD patients,20 while Xun et al. similarly found an increased prevalence of Veillonella coupled with decreased Prevotella, Neisseria and Haemophilus.21 Furthermore, when comparing the salivary CD patients in remission to active CD, Zhang et al. found reduced Neisseria, Haemophilus, Fusobacterium and Porphyromonas in active CD.22 Although the exact characterization differed between the studies, they all found that oral microbiome dysbiosis occurs in CD. As a diagnostic tool, the oral microbiome shows great promise as saliva samples are non-invasive unlike colonoscopy, and are easy to collect at any time unlike faecal samples. However, the lack of a comprehensive characterization, taking into account conditions that can affect the oral microbiome such as dental caries, periodontitis and oral manifestations,23 limits the usefulness of oral microbiome diagnostics in CD.
In this study, all participants underwent oral examination by a board-certified dentist, prior to oral microbiome sampling. Additionally, potential confounders such as the presence of dental diseases and oral manifestations were considered in the characterization. This study characterized the oral microbiome of CD patients, with and without oral manifestations, compared to healthy controls with the aim to identify the association between compositional and functional alterations in the oral manifestation of CD using shotgun metagenomics sequencing in a mixed population consisting of Han Chinese, Malay, Indian and Caucasian subjects.
2. Materials and Methods
2.1. Clinical cohort
This cross-sectional study enrolled patients at the National University Hospital [NUH], Singapore, between May 2017 and January 2019. Eligible patients needed to be 21 years of age or older, and to either have an established diagnosis or exclusion of CD. Patients currently on antibiotics or with recent exposure to probiotics/antibiotics within a month, or with significant comorbidities such as active cancer were excluded. Patients with CD [with and without oral manifestations] were recruited from the IBD subspeciality clinic under the Division of Gastroenterology, NUH. Non-CD healthy controls were age, race and gender matched with CD subjects without oral manifestations and recruited from the Dental Center, NUH University Dental Cluster. They were also screened prior to enrolment and required to have no known gastrointestinal signs and symptoms such as diarrhoea, abdominal pain and blood in the stool, as well as no known concomitant systematic diseases. Upon providing informed consent, patients completed detailed questionnaires, which collected information on demographics, history of tobacco use, other comorbidities, current medications, as well as self-reported gastrointestinal symptoms such as diarrhoea, abdominal pain and blood in the stool. Patients with CD had their disease activity scored according to the Crohn’s Disease activity index [CDAI].24 All experiments were performed in accordance with all relevant regulations and guidelines. The study received ethics approval from the institutional review boards of the National Healthcare Group, Singapore [DSRB reference E/2016/01285].
2.2. Assessment of oral condition
All subjects underwent an oral examination by a single dentist [S.H.] conducted under artificial light. Dental conditions known to affect the oral microbiome such as oral hygiene, caries and periodontal disease were recorded. Additionally, the presence and type of oral manifestations such as mucogingivitis, mucosal tags, deep ulceration, cobblestoning and lip swelling were recorded during the examination of CD patients. This was supplemented by eliciting any history of oral manifestations.
2.3. Microbiome sample collection
The oral microbiome was sampled through participants self-collection of saliva. Participants were instructed in person, and supplemented with visual aids such as videos and hand-out pamphlets, on how to collect their own saliva specimens. Participants were explicitly instructed to refrain from eating [including chewing gum and sweets], smoking or dental procedures [including toothbrushing and mouthwash use] for 1 h before saliva collection. Saliva was collected using the OMNIgene Discover Kit 505 [DNA Genotek], according to the manufacturer’s instructions. All specimens were collected on the day of examination. The saliva samples were stored at room temperature as per the manufacturer’s recommendation and sent for DNA extraction within 4 weeks of collection.
2.4. Microbiome profiling
DNA was extracted from saliva samples after mechanical lysis via bead-beating, using Exgene Clinic SV Mini kits [GeneAll Biotechnology] according to the manufacturer’s instructions. Extracted DNA was purified using AMPure XP beads [Beckman Coulter]. The quantity and quality of DNA were examined using a NanoDrop 8000 Spectrophotometer [Thermo Fisher Scientific]. Extracted DNA samples were stored at −80°C and sent for library preparation within 6 weeks of extraction. Indexed sequencing libraries were prepared using a QIAGEN QIAseq FX DNA Library Kit [Qiagen] following the manufacturer’s instructions and sequenced as 2 × 151 bp paired-end reads on an Illumina HiSeq 4000 sequencer [Illumina]. On average, 16.6 ± 12.1 million raw read pairs were obtained, with 14.1 ± 10.5 million obtained after quality control for each sample. The average host reads were 11.6 ± 9.9 million, and after decontamination 2.1 ± 3.2 million microbial read pairs were present.
Metagenomic taxonomic and functional profiles were generated using the bioBakery meta’omics workflow25,26 in the Terra workspace. Briefly, reads mapping to the human genome were first filtered out using KneadData. Taxonomic profiles of shotgun metagenomes were generated using MetaPhlan3, which uses a library of clade-specific markers to provide pan-microbial profiling. Functional profiling was performed by HUMAnN3, whereby HUMAnN3 constructs a sample-specific reference database from the pangenomes of the subset of species detected in the samples by MetaPhlAn3 [pangenomes are precomputed representatives of the open reading frames of a given species]. Sample reads are mapped against this database to quantify gene presence and abundance on a per-species basis. A translated search is then performed against a UniRef-based protein sequence catalogue [UniRef release 2020] for all reads that fail to map at the nucleotide level. The results are abundance profiles of gene families [UniRef90s], for both metagenomics, stratified by each species contributing to those genres, and further summarized to higher level gene groups such as Enzyme Commission (EC), KEGG Orthology (KO) or metaCYC pathways.
For subsequent analysis, read counts were transformed into relative abundances by normalization to the total number of reads per sample. Low-abundance filters were applied to discard taxonomic and functional features whose relative abundance did not reach 0.1% and 0.001% respectively, in at least 10% of the individuals.
2.5. Statistical analysis
Alpha diversity [richness and Shannon diversity] was calculated for taxonomic profiles at the species level, using the Vegan package in R.27 Linear models were used to identify putative differential abundance analysis of metagenomic features, with fitting performed with the MaAsLin2 package in R,28 with nominal p-values adjusted for multiple hypothesis testing with a target false discovery rate (FDR) of 0.2. In brief, metagenomic features were log transformed to variance-stabilize the data. Zero values were additively smoothed by half of the minimal abundance for each feature and then fitted with the following per-feature linear mixed-effect model:
In each per-feature multivariable model, the transformed abundance of each feature was modelled as a function of oral lesions nested within binary variables [with no lesions as the reference] with the presence or absence of CD, while adjusting for activity [no IBD activity as reference], duration of disease [continuous variable], sequencing depth [continuous variable], and concomitant steroid and azathioprine use [both binary covariates].
To construct a prediction model for CD diagnosis using saliva samples, relative abundances of microbial species were first converted to log10 relative abundances, then z-normalized, before passing through a random forest [RF] classifier to distinguish subjects with and without CD. The performance of the RF classifier was evaluated using five-fold cross validation and generation of a summary receiver operating characteristic (ROC) curve with 95% confidence interval for the classifier result. The five-fold testing model was then combined with a model build of 2000 trees and mtry grid search of 1–150 variables using the R package Caret.29
2.6. Data availability
Whole-genome sequencing data that support the findings of this study are available in the European Nucleotide Archive with the primary accession code PRJEB39813.
3. Results
3.1 Study population
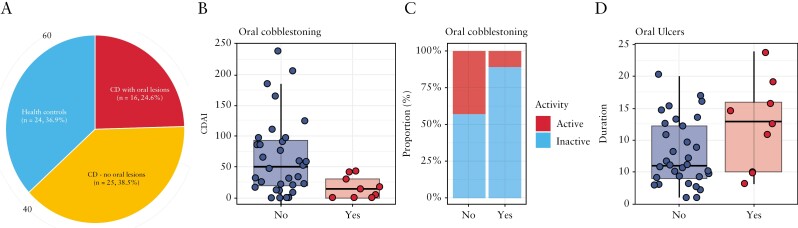
The study recruited a total of 65 subjects, with a mean age of 39.5 [range: 21–67] years [Table 1]. One-third of the study population [n = 24, 36.9%] were non-CD healthy controls [Figure 1a]. Amongst the 41 subjects with CD, two-fifths had oral manifestations [n = 16, 39.0%], of whom patients had either oral ulcers [n = 7, 44.4%], cobblestoning [n = 7, 44.4%] or both [n = 2]. Other reported oral lesions included mucogingivitis [n = 4], mucosal tags [n = 2] and lip swelling [n = 1]. Subjects with CD were similar to controls in demographic covariates including age, gender and smoking history. However, subjects with oral lesions were more likely to be male [81.2% vs 44.0%, p < 0.05] and have the concomitant usage of azathioprine [81.2% vs 48.0%, p = 0.07]. Of note, subjects with cobblestoning had much lower CDAI scores [mean 65.2 vs 16.8, p = 0.03, Figure 1b], which was consistent with a lower proportion of clinically active disease [43.8% vs 11.1%, Figure 1c]. Subjects with oral ulcers reported no significant differences in CDAI and clinical activity, although subjects with oral ulcers were found to have had CD for much longer periods of time [12.3 vs 7.9 years, p = 0.04, Figure 1d].
Table 1.
Demographics, and clinical and oral conditions of subjects [n = 65]
| Crohn’s disease, oral manifestation absent [CDA], n = 25 | Crohn’s disease, oral manifestation present [CDP], n = 16 | Healthy controls [HC], n = 24 | p-value | |
|---|---|---|---|---|
| Demographics | ||||
| Mean age, years [SD] | 39.1 [14.4] | 38.6 [12.0] | 40.2 [1] | 0.84 |
| Ethnicity, n [%] | ||||
| Chinese | 12 [48.0] | 10 [62.5] | 12 [50.0] | 0.948 |
| Malay | 3 [12.0] | 2 [12.5] | 3 [12.5] | |
| Indian | 9 [36.0] | 3 [18.8] | 9 [37.5] | |
| Caucasian | 1 [4.0] | 1 [6.2] | 0 [0.0] | |
| Sex, n [%] | ||||
| Male | 11 [41.7] | 13 [81.2] | 10 [41.7] | 0.028* |
| Female | 14 [58.3] | 3 [18.8] | 14 [58.3] | |
| Smoking, n [%] | ||||
| Non-smoker | 20 [83.3] | 13 [81.2] | 20 [80.0] | 0.913 |
| Previous/current smoker | 5 [16.7] | 3 [18.8] | 4 [20.0] | |
| Crohn’s disease condition | ||||
| Duration [SD] | 7.9 [5.5] | 10.4 [6.2] | 0.181 | |
| Crohn’s disease activity index [SD] | 61.5 [58.4] | 43.8 [62.1] | 0.361 | |
| Activity | 10 [40.0] | 5 [31.2] | 0.814 | |
| C-reactive protein level [CRP] [mg/L], mean [SD] | 8.8 [17.5] | 6.9 [10.6] | 0.713 | |
| Location, n [%] | 0.952 | |||
| L1 | 2 [8.0] | 1 [6.2] | ||
| L2 | 7 [28.0] | 5 [31.2] | ||
| L3 | 14 [56.0] | 8 [50.0] | ||
| L4 | 2 [8.0] | 2 [12.5] | ||
| Behaviour, n [%] | 0.890 | |||
| B1 | 13 [52.0] | 9 [56.2] | ||
| B2 | 1 [4.0] | 1 [6.2] | ||
| B3 | 11 [44.0] | 6 [37.5] | ||
| Perianal involvement, n [%] | 0.898 | |||
| No | 17 [68.0] | 12 [75.0] | ||
| Yes | 8 [32.0] | 4 [25.0] | ||
| Age of CD onset, n [%] | 0.467 | |||
| A1 | 3 [12.0] | 1 [6.2] | ||
| A2 | 14 [56.0] | 12 [75.0] | ||
| A3 | 8 [32.0] | 3 [18.8] | ||
| Medication † , n [%] | ||||
| Steroids/anti-inflammatory [mesalazine] | 5 [20.0] | 3 [18.8] | 0.453 | |
| Azathioprine | 12 [48.0] | 13 [81.2] | 0.072 | |
| Biologics | 10 [40.0] | 10 [62.5] | 0.278 | |
| Others [tacrolimus, sulphasalazine] | 2 [8.0] | 2 [12.5] | 0.224 | |
| Dental condition, n | ||||
| Presence of decayed teeth [%] | 22 [88.0] | 12 [75.0] | 18 [75.0] | 0.444 |
| DMFT: severity of decayed teeth [SD] | 6.1 [7.0] | 3.8 [3.9] | 4.7 [5.3] | 0.427 |
| Periodontal condition: presence of periodontitis [%] | 5 [20.8] | 4 [25.0] | 5 [20.8] | 0.925 |
| Type of oral manifestation ‡, n [%] | ||||
| Cobblestoning | 8 [50.0] | |||
| Lip swelling | 1 [6.5] | |||
| Mucogingivitis | 4 [25.0] | |||
| Mucosal Tags | 3 [18.8] | |||
| Ulcers | 10 [62.5] | |||
p < 0.05.
Some subjects were on multiple drug regimens.
Some subjects had multiple types of oral manifestations.
Figure 1.
Crohn’s disease profile and its relationship with oral manifestations. [a] Distribution of subjects based on presence of IBD [Crohn’s disease] and oral manifestations. [b] Crohn’s disease activity index [CDAI] scores of CD subjects with and without oral cobblestoning. [c] Disease activity of CD subjects with and without oral cobblestoning. [d] Duration of disease of CD subjects with and without oral ulcers.
3.2. Oral microbiome dysbiosis occurs in CD and variance is determined by four key variables
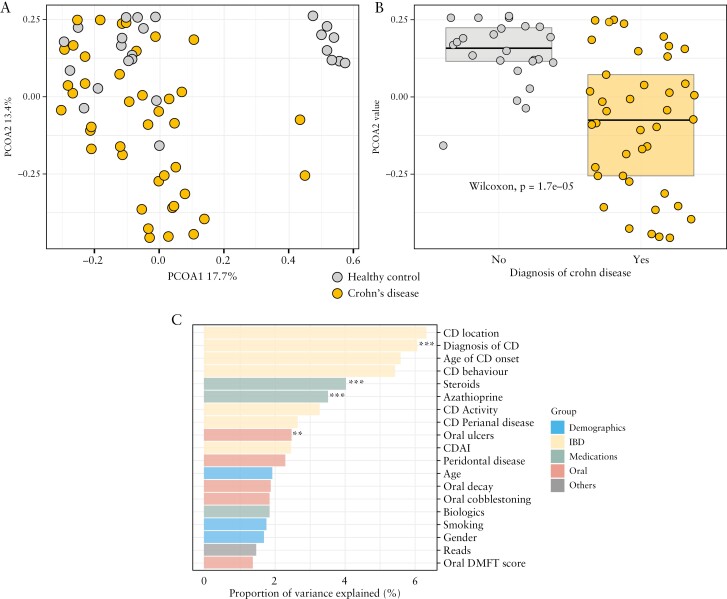
Major variations in the subjects’ taxonomic profiles were driven by gradients of Bacteroidota [Bacteroidetes] and Bacillota [Firmicutes] relative abundances across the cohort [Supplementary Figure 1], which corresponded significantly to a history of CD. Patients with CD were found to have increased saliva microbial species of the phyla Actinomycetota [Actinobacteria] and Pseudomonadota [Proteobacteria], but no significant differences in the total abundance of microbial species of the phyla Bacillota [Firmicutes] and Bacteroidota [Bacteroidetes], with a corresponding borderline increase in the ratio of Bacillota [Firmicutes] to Bacteroidota [Bacteroidetes], though not statistically significant [p = 0.44] [Supplementary Figure 2]. Unsupervised principal coordinate analysis [PCoA] conducted on Bray–Curtis dissimilarity matrices [Figure 2a] demonstrated distinct oral microbiome profiles between subjects with and without CD. Subjects with CD had significantly lower PCoA2 [p < 0.04, Figure 2b], whereby PCoA1 and PCoA2 represented 17.7% and 13.4%, respectively, of the overall microbiome variance.
Figure 2.
Crohn’s disease is a major determinant of oral microbiome structure. [a] Unsupervised principal coordinate analysis [PCoA] conducted on Bray–Curtis dissimilarity matrices demonstrating distinct oral microbiome profiles between subjects with and without CD. [b] PCoA2 score is significantly [p < 0.05] different between subjects with and without CD. [c] PERMANOVA analysis showing four key variables [**p < 0.1, ***p < 0.05] explaining overall oral microbiome community variance.
To further identify significant associations between overall oral microbiome structure with host demographics, CD clinical variables and oral hygiene, a permutational multivariate analysis of variance (PERMANOVA)-based testing utilizing Bray–Curtis dissimilarity profiles found four key variables: [1] diagnosis of CD, [2] concomitant use of steroids, [3] concomitant use of azathioprine and [4] presence of oral ulcers. These variables explained 6.1, 4.0, 3.5 and 2.5% of the overall oral microbiome community variance [Figure 2c]. Dental conditions such as the presence of periodontitis and dental caries accounted for up to 3% of the variation in overall salivary microbiome composition, though this was not statistically significant [p > 0.05].
There were no differences in alpha diversity found among the groups [Supplementary Figure 3].
3.3. Differences in oral microbiome composition at the species level
When examining the taxonomic proportion of the three main study groups [healthy controls, CD with oral lesions, CD without oral lesions], significant differences were found between subjects with and without CD as well as amongst CD subjects with and without oral lesions [Supplementary Figure 4].
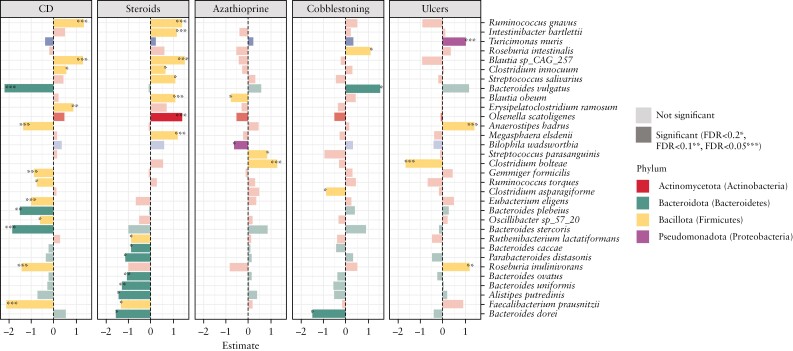
Next, the specific taxonomic signatures were explored at the species level, associated with either of the two predominant oral manifestations [i.e. cobblestoning, oral ulcers]. This was conducted using multivariable linear models, while correcting for the concomitant use of steroids or azathioprine, and nesting for the diagnosis of CD. A total of 32 significant differentially abundant microbial species were identified, of which the majority were associated with the diagnosis of CD [n = 14] or the concomitant use of steroids [n = 16] [Figure 3]. Four microbial species each were found associated with either cobblestoning [Roseburia intestinalis, Bacteroides vulgatus, Clostridium asparagiforme and Bacteriodes dorei] or oral ulcers [Turicimonas muris, Anaerostipes hadrus, Clostridium bolteae and Roseburia inulinivorans].
Figure 3.
Significant differentially abundant microbial species with respect to the key variables and oral manifestations [*FDR < 0.2, **FDR < 0.1, ***FDR < 0.05].
3.4. Functional differences of the oral microbiome in CD subjects with oral ulcers
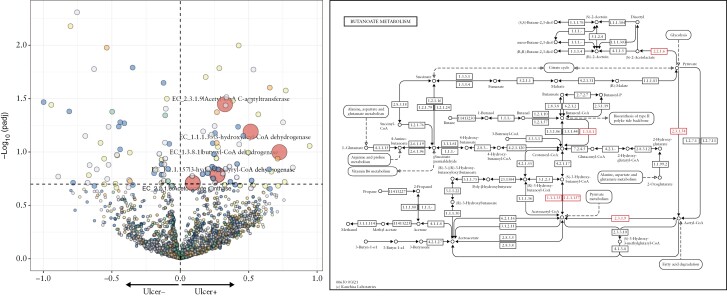
To understand the functional differences due to microbial composition differences in CD subjects with oral manifestations, a differential abundance analysis of the gene families in the metagenome was conducted. The relative abundances were summed according to their EC annotations, using the same linear modelling approach described above. A total of 676 differentially abundant microbial enzymes were identified, of which the majority were associated with the diagnosis of CD [n = 455], concomitant use of steroids [n = 111] and presence of oral ulcers [n = 105] [Supplementary Table S1]. Focusing on oral ulcers, the majority of these differently abundant enzymes were associated with microbial metabolism in a diverse environment [n = 25], involved in biosynthesis of secondary metabolism [n = 16] and metabolism of amino acids such as phenylalanine [n = 6], valine, leucine and isoleucine [n = 4], when binned to their higher Kyoto Encyclopedia of Genes Genomes modules. Of interest, an increase of microbial enzymes [n = 5] associated with butyrate metabolism [Figure 4] was found to be positively associated with the presence of oral ulcers. Specifically, the acetyl-CoA-based pathway for butyrate production was found to be enriched.
Figure 4.
Functional analysis of microbial genes in subjects with and without ulcer ulcers. [a] Volcano plot of microbial enzymes associated with and without oral ulcers. The five enzymes on the upper right quadrant are associated with increased butyrate production. [b] The butanoate [butyrate] metabolism pathway with enzymes [highlighted red] associated with the presence of oral ulcers.
3.5 Oral [salivary] microbes can aid in the diagnosis of CD
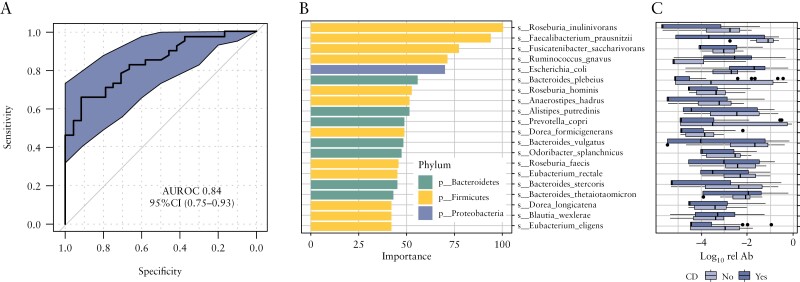
Several studies have explored the potential for diagnosing IBD based on microbial composition, using stool30 or intestinal mucosal samples.5 The current study evaluated how microbiome composition in the oral cavity through collection of saliva performs for classifying subjects with and without CD using ROC analysis. Using a five-fold cross validation RF classifier, we found that the oral microbiome has strong discriminatory function [accuracy: 74%] to distinguish subjects with and without CD [AUROC 0.84, Figure 5a]. The top contributory microbial species [Figure 5b and c] mirrored the prominent differentially abundant stool microbial species associated with IBD.
Figure 5.
Model of the oral microbiome predicting the presence of CD. [a] Receiver operating characteristic [ROC] analysis [AUROC: 0.84]. [b] Differentially abundant oral microbial species selected for the model. [c] Relative abundance of oral microbial species in the model.
4. Discussion
The present study found that the salivary oral microbiome can be used in a predictive model for the presence of CD with an accuracy over 70%. This opens up the possibility of using a diagnostic test that is non-invasive and easy to conduct, compared to the traditional investigations in CD such as colonoscopy, which is invasive and exposes the patient to the risks of sedation; or compared to stool tests, which can be an uncomfortable prospect for patients to provide.31 Although much work had been done in using the microbiome as a diagnostic and prognostic tool in IBD, most of it has been focused on the gut microbiome.32 A recent study examined the use of the gut microbiome as a diagnostic tool for CD and reported an overall sensitivity of 80% for the detection of CD,2 while a study using 16S rRNA gene sequencing was able to differentiate IBD subjects from healthy controls at over 70% using the salivary oral microbiome.33 These were comparable to the present study, with the present study having the advantage of characterizing saliva samples with the use of whole genome sequencing over 16S sequencing.
This study also found that key factors contributing to the oral microbiome dysbiosis in CD were related to underlying CD, with local oral diseases such as periodontitis and dental caries having smaller contributions. As such, the oral microbiome profiled using saliva samples may aid in the diagnosis of CD with minimal interference from concurrent oral diseases. Differentially abundant oral microbes in CD were predominantly the same stool species associated with IBD [i.e. increased abundances of Ruminococcus gnavus, decreased abundance of Faecalibacterium prausnitzii].34 Surprisingly, there were very few oral commensals that were part of the predictive model, which could be the result of the oral microbiome dysbiosis present in CD subjects. This was not the case when other types of oral samples were collected, as illustrated by plaque microbiome dysbiosis detected in periodontitis35 and dental caries.36 The other two key factors could be related to the severity of disease as the use of steroids and azathioprine are typically reserved for the management of more severe disease.37 The microbial species associated with concomitant steroid usage largely overlap with the significant microbial species associated with CD. This suggests that the use of steroids can further tip the oral microbiome community of CD subjects towards a dysbiotic state, similar to previous findings by Gevers and colleagues who reported a similar phenomenon seen with antibiotic treatment in CD subjects.5 However, this finding should be interpreted with caution, as are many steroid-associated confounders.
Additionally, this was the first study to profile the oral microbiome in association with the oral manifestations of CD. It was found that the presence of oral ulcers was a key factor in the composition of the oral microbiome. There are existing studies linking the gut microbiome composition to the activity of CD38 as well as predicting the severity of intestinal inflammation in IBD,39 potentially allowing its use as a prognostic tool in the management of CD. This study found that some oral manifestations were associated with disease activity [cobblestoning] and duration of CD [oral ulcers]. Four species were identified to be associated with cobblestoning and another group of four species were associated with oral ulcers in the present study. Although currently insufficient to provide an accurate model, it provides a starting point for future studies to explore the use of the oral microbiome as a prognostic test in CD.
Around 40% of the CD subjects presented with oral manifestations, which was similar to previous studies.14 Previous research in murine models had shown that the disease only manifests in susceptible genotypes and is driven by microbial dysbiosis.40 The entire gastrointestinal tract, including the oral cavity, can be involved in CD, and inflammatory lesions were postulated to be caused by impaired interactions between the commensal microbiome with the human host. The present study showed that oral microbiome dysbiosis was associated with oral manifestations, such as cobblestoning and oral ulcers, found in CD subjects. There have been attempts to alter the microbiome using targeted antimicrobial approaches such as the use of antibiotics, prebiotics and probiotics41; however, their use in CD has so far been hampered by the failure to define a core gut microbiome, as well as difficulty in delivering therapeutics to the gut.42 The oral cavity is much more accessible than the rest of the alimentary canal and can be a useful model for the development of new treatment targeting microbiome dysbiosis.
Additionally, differences in the functional pathways appear to support the hypothesis of oral microbiome dysbiosis resulting in the development of oral manifestations in CD subjects. Of the four known substrate pathways of butyrate production, the acetyl-CoA pathway was found to be enriched in the present study. This pathway is associated with the fermentation of polysaccharides from the diet,43 a potential factor in the enrichment of butyrate in these subjects. There are many butyrogenic bacteria in the oral microbiome, and there was evidence to suggest that butyrate generated by periodontal pathogens during metabolism plays a considerable role in the initiation and progression of periodontitis.44 Butyrate was found to increase inflammation in periodontal tissues and causes reactivation of latent viruses, such that the increase of butyrate can be a factor in the development of oral manifestations in CD. This was further substantiated by the finding of an increased abundance of A. hadrus in subjects with oral ulcers, a major butyrate producer usually found in the gut.45 On the other hand, the protective mechanisms of butyrate in IBD have been well documented in the colon, from decreasing inflammation to increasing mucus production.46 Therapies centred around butyrate have been developed, such as supplementation of butyrate-producing bacteria in the gut microbiome47 and the use of oral butyrate to manage symptoms.48 This suggests that the mechanism of inflammatory destruction may differ between the oral and gut environment.
One of the limitations of this study is the reliance on clinical examination to determine the presence of oral manifestations, which may result in missing some subjects due to the cyclical nature of CD. Moreover, a clinical index [CDAI] was used to measure current disease activity, which may have issues with accuracy. Future studies should consider the use of more objective measures such as faecal calprotectin levels and endoscopic assessment to measure current disease activity. Additionally, the relatively small sample size and cross-sectional nature of the study may limit its statistical power. Lastly, diet differences have been known to affect the composition of the oral microbiome49; this was not explored in the present study and should be considered in future studies. Despite the study limitations, there were several important and novel findings. First, a predictive model for the presence of CD had comparable accuracy to models utilizing the gut microbiome, therefore providing the possibility of using saliva as a diagnostic tool. Second, groups of salivary microbes were found to be associated with the development of oral manifestations in CD. Lastly, it highlighted the importance of the butyrate pathway in the oral manifestation in CD. These findings demonstrated the importance of the oral microbiome in CD and the opportunity for it to be used in the diagnosis and management of CD.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Paola de Sessions, Dr Eileen Png and Ms Song Jie for their contribution to the microbial DNA extraction and sequencing. The authors are also grateful to Prof. Ramnik J. Xavier for helpful discussions, and comments on the manuscript.
Contributor Information
Shijia Hu, Faculty of Dentistry, National University of Singapore, Singapore.
John Mok, Division of Gastroenterology & Hepatology, National University Hospital, Singapore.
Michelle Gowans, Division of Gastroenterology & Hepatology, National University Hospital, Singapore.
David E H Ong, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Juanda Leo Hartono, Division of Gastroenterology & Hepatology, National University Hospital, Singapore; Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Jonathan Wei Jie Lee, Division of Gastroenterology & Hepatology, National University Hospital, Singapore; Yong Loo Lin School of Medicine, National University of Singapore, Singapore; Broad Institute of MIT and Harvard, Cambridge, MA, USA; NUS Synthetic Biology for Clinical and Technological Innovation (SynCTI), Singapore.
Funding
This work was supported by the National University of Singapore Start-up grant [Exploring the Oral Microbiome in Crohn’s Disease] [A-0002940-00-0].
Conflict of Interest
J.L.W.J. and D.E.H.O. are co-founders of AMILI and serve as members of the scientific advisory board. The other authors have no conflicts of interest to declare.
Author Contributions
S.H. conceived the idea; S.H., J.M., M.G., J.L.H. and D.E.H.O. participated in collecting the samples; J.L.W.J. conducted the data analysis; S.H. and J.L.W.J. led the writing; J.M., M.G., J.L.H. and D.E.H.O. revised the manuscript for important intellectual content.
References
- 1. Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L.. Crohn’s disease. Lancet 2017;389:1741–55. [DOI] [PubMed] [Google Scholar]
- 2. Pascal V, Pozuelo M, Borruel N, et al. A microbial signature for Crohn’s disease. Gut 2017;66:813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007;2:119–29. [DOI] [PubMed] [Google Scholar]
- 4. Frank DN, Robertson CE, Hamm CM, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 2011;17:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elmaghrawy K, Hussey S, Moran GP.. The oral microbiome in pediatric IBD: a source of pathobionts or biomarkers? Front Pediatr 2020;8:620254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu S, Png E, Gowans M, et al. Ectopic gut colonization: a metagenomic study of the oral and gut microbiome in Crohn’s disease. Gut Pathog 2021;13:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Read E, Curtis MA, Neves JF.. The role of oral bacteria in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2021;18:731–42. [DOI] [PubMed] [Google Scholar]
- 10. She YY, Kong XB, Ge YP, et al. Periodontitis and inflammatory bowel disease: a meta-analysis. BMC Oral Health 2020;20:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graham DB, Xavier RJ.. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020;578:527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baumgart DC, Sandborn WJ.. Crohn’s disease. Lancet 2012;380:1590–605. [DOI] [PubMed] [Google Scholar]
- 13. Vavricka SR, Schoepfer A, Scharl M, et al. Extraintestinal manifestations of inflammatory bowel disease. Inflamm Bowel Dis 2015;21:1982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lankarani KB, Sivandzadeh GR, Hassanpour S.. Oral manifestation in inflammatory bowel disease: a review. World J Gastroenterol 2013;19:8571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harty S, Fleming P, Rowland M, et al. A prospective study of the oral manifestations of Crohn’s disease. Clin Gastroenterol Hepatol 2005;3:886–91. [DOI] [PubMed] [Google Scholar]
- 16. Eckel A, Lee D, Deutsch G, Maxin A, Oda D.. Oral manifestations as the first presenting sign of Crohn’s disease in a pediatric patient. J Clin Exp Dent 2017;9:e934–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Docktor MJ, Paster BJ, Abramowicz S, et al. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm Bowel Dis 2012;18:935–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelsen J, Bittinger K, Pauly-Hubbard H, et al. Alterations of the subgingival microbiota in pediatric Crohn’s disease studied longitudinally in discovery and validation cohorts. Inflamm Bowel Dis 2015;21:2797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Brien CL, Kiely CJ, Pavli P.. The microbiome of Crohn’s disease aphthous ulcers. Gut Pathogens 2018;10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Said HS, Suda W, Nakagome S, et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res 2014;21:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xun Z, Zhang Q, Xu T, Chen N, Chen F.. Dysbiosis and ecotypes of the salivary microbiome associated with inflammatory bowel diseases and the assistance in diagnosis of diseases using oral bacterial profiles. Front Microbiol 2018;9:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang T, Kayani M, Hong L, et al. Dynamics of the salivary microbiome during different phases of Crohn’s disease. Front Cell Infect Microbiol 2020;10:544704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sampaio-Maia B, Caldas I, Pereira M, Pérez-Mongiovi D, Araujo R.. The oral microbiome in health and its implication in oral and systemic diseases. Adv Appl Microbiol 2016;97:171–210. [DOI] [PubMed] [Google Scholar]
- 24. Best WR, Becktel JM, Singleton JW, Kern F.. Development of a Crohn’s disease activity index. National Cooperative Crohn’s Disease Study. Gastroenterology 1976;70:439–44. [PubMed] [Google Scholar]
- 25. Beghini F, McIver LJ, Blanco-Míguez A, et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with biobakery 3. Elife 2021;10:e65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McIver LJ, Abu-Ali G, Franzosa EA, et al. Biobakery: a meta’omic analysis environment. Bioinformatics 2018;34:1235–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oksanen J, Kindt R, Legendre P, et al. The vegan package. Commun Ecol Packag 2007;10:719. [Google Scholar]
- 28. Mallick H, Rahnavard A, McIver LJ, et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput Biol 2021;17:e1009442e1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhn M. Caret: classification and regression training. Astrophysics Source Code Library 2015:ascl: 1505.003. [Google Scholar]
- 30. Papa E, Docktor M, Smillie C, et al. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 2012;7:e39242e39242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laass MW, Roggenbuck D, Conrad K.. Diagnosis and classification of Crohn’s disease. Autoimmun Rev 2014;13:467–71. [DOI] [PubMed] [Google Scholar]
- 32. Aldars-García L, Chaparro M, Gisbert JP.. Systematic review: the gut microbiome and its potential clinical application in inflammatory bowel disease. Microorganisms 2021;9:977977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Somineni HK, Weitzner JH, Venkateswaran S, et al. Site-and taxa-specific disease-associated oral microbial structures distinguish inflammatory bowel diseases. Inflamm Bowel Dis 2021;27: 1889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schirmer M, Garner A, Vlamakis H, Xavier RJ.. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 2019;17:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Curtis MA, Diaz PI, Van Dyke TE.. The role of the microbiota in periodontal disease. Periodontology 2000 2020;83:14–25. [DOI] [PubMed] [Google Scholar]
- 36. Tanner A, Kressirer C, Rothmiller S, Johansson I, Chalmers N.. The caries microbiome: implications for reversing dysbiosis. Adv Dent Res 2018;29:78–85. [DOI] [PubMed] [Google Scholar]
- 37. Lichtenstein GR, Hanauer SB, Sandborn WJ; Gastroenterology PPCotACo. Management of Crohn’s disease in adults. Off J Am Coll Gastroenterol ACG 2009;104:465–83. [DOI] [PubMed] [Google Scholar]
- 38. Magro DO, Santos A, Guadagnini D, et al. Remission in Crohn’s disease is accompanied by alterations in the gut microbiota and mucins production. Sci Rep 2019;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Malham M, Lilje B, Houen G, et al. The microbiome reflects diagnosis and predicts disease severity in paediatric onset inflammatory bowel disease. Scand J Gastroenterol 2019;54:969–75. [DOI] [PubMed] [Google Scholar]
- 40. Bloom SM, Bijanki VN, Nava GM, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe 2011;9:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Preidis GA, Versalovic J.. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: Gastroenterology enters the metagenomics era. Gastroenterology 2009;136:2015–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Khan I, Ullah N, Zha L, et al. Alteration of gut microbiota in inflammatory bowel disease (IBD): Cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 2019;8:126126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vital M, Howe AC, Tiedje JM.. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. MBio 2014;5:e00889–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guan X, Li W, Meng H.. A double-edged sword: role of butyrate in the oral cavity and the gut. Mol Oral Microbiol 2021;36:121–31. [DOI] [PubMed] [Google Scholar]
- 45. Kant R, Rasinkangas P, Satokari R, Pietilä TE, Palva A.. Genome sequence of the butyrate-producing anaerobic bacterium Anaerostipes hadrus pel 85. Genome Announc 2015;3:e00224–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Silva JP, Navegantes-Lima KC, Oliveira AL, et al. Protective mechanisms of butyrate on inflammatory bowel disease. Curr Pharm Des 2018;24:4154–66. [DOI] [PubMed] [Google Scholar]
- 47. Geirnaert A, Calatayud M, Grootaert C, et al. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci Rep 2017;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sabatino AD, Morera R, Ciccocioppo R, et al. Oral butyrate for mildly to moderately active Crohn’s disease. Aliment Pharmacol Ther 2005;22:789–94. [DOI] [PubMed] [Google Scholar]
- 49. Kato I, Vasquez A, Moyerbrailean G, et al. Nutritional correlates of human oral microbiome. J Am Coll Nutr 2017;36:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.