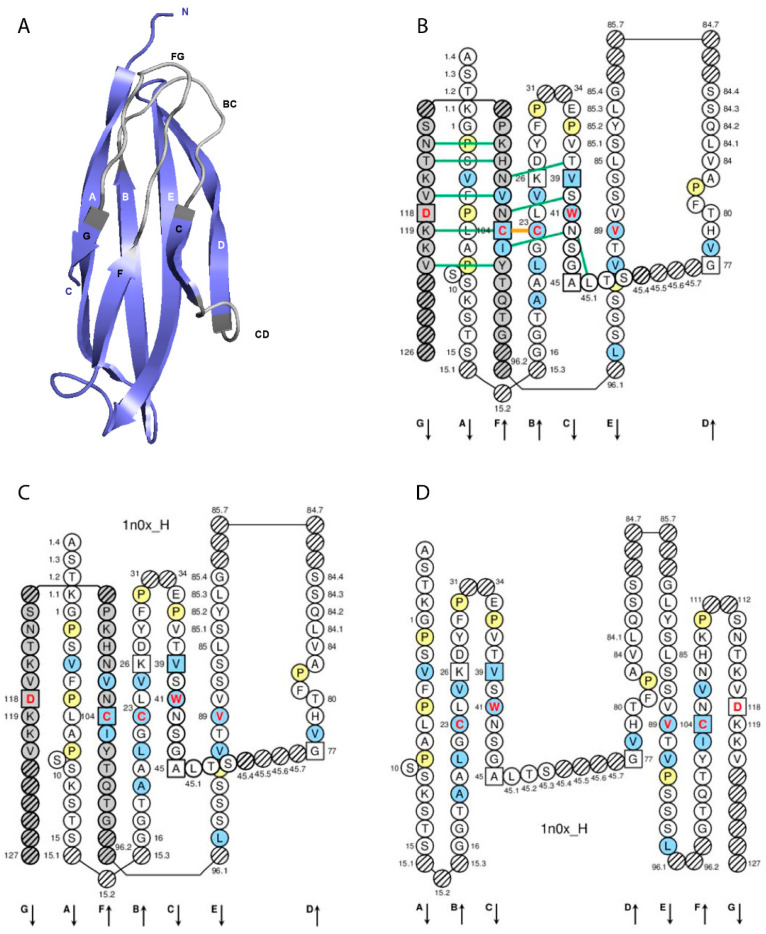
Figure 3.
IG constant (C) domain. (A) 3D structure ribbon representation with the IMGT strand and loop delimitations. (B) IMGT Collier de Perles on two layers with hydrogen bonds. The IMGT Colliers de Perles on two layers show, in the forefront, the GFC strands, and in the back, the ABED strands (located at the interface CH1/CL of the IG), linked by the CD transversal strand. The IMGT Collier de Perles with hydrogen bonds (green lines online, only shown here for the GFC sheet) is generated by the IMGT/Collier de Perles tool [51] integrated in the IMGT/3Dstructure-DB, from experimental 3D structure data. (C) IMGT Collier de Perles on two layers from IMGT/DomainGapAlign [28,49,50]. (D) IMGT Colliers de Perles on one layer. Amino acids are shown in the one-letter abbreviation. All proline (P) are shown online in yellow. IMGT anchors are represented by squares. Hatched circles are IMGT gaps according to the IMGT unique numbering for the C-domain [68]. Positions with bold (online red) letters indicate the four conserved positions that are common to a V-domain and to a C-domain: 23 (1st-CYS), 41 (CONSERVED-TRP), 89 (hydrophobic), 104 (2nd-CYS), and position 118, which is only conserved in V-DOMAIN. The identifier of the chain to which the CH-domain belongs is 1n0x_H (from the Homo sapiens b12 Fab, in IMGT/3Dstructure-DB, https://www.imgt.org) [27,28,29]. The 3D ribbon representation was obtained using PyMOL and “IMGT numbering comparison” of 1n0x_H (CH1) from IMGT/3Dstructure-DB (https://www.imgt.org) [27,28,29].