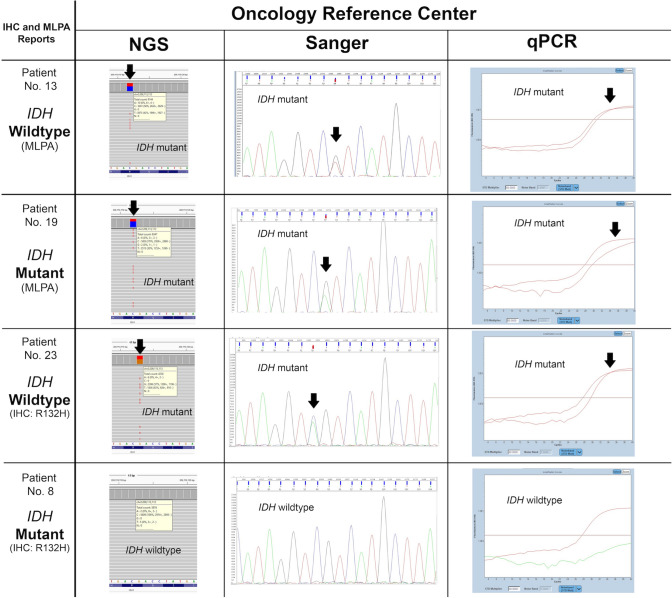
Fig. 6.
Examples of IHC and MLPA reports from different non-oncological reference centers versus NGS, qPCR, and Sanger sequencing results from the oncology reference center. Two false positives were detected by MLPA and IHC, and one false negative by IHC. Pathogenic variants detected by NGS were additionally validated by two independent methods: Sanger sequencing and qPCR. The black arrows indicate mutations detected using next-generation sequencing (NGS Targeted Hotspot Panel, Entrogen), Sanger sequencing (Applied Biosystems SeqStudio Genetic Analyzer), and qPCR (IDH1/2 Mutation Detection Kit, Entrogen), with a limit of detection of 5, 20, and 1%, respectively. Two red curves represent (1) positive control and (2) amplicon with mutation detected (arrow). The patient no. 13 – MLPA report indicates IDH-wildtype, while NGS, qPCR, and Sanger sequencing confirmed mutation in the IDH1 gene. IHC immunohistochemistry, MLPA multiplex ligation-dependent probe amplification, NGS next-generation sequencing, qPCR quantitative polymerase chain reaction