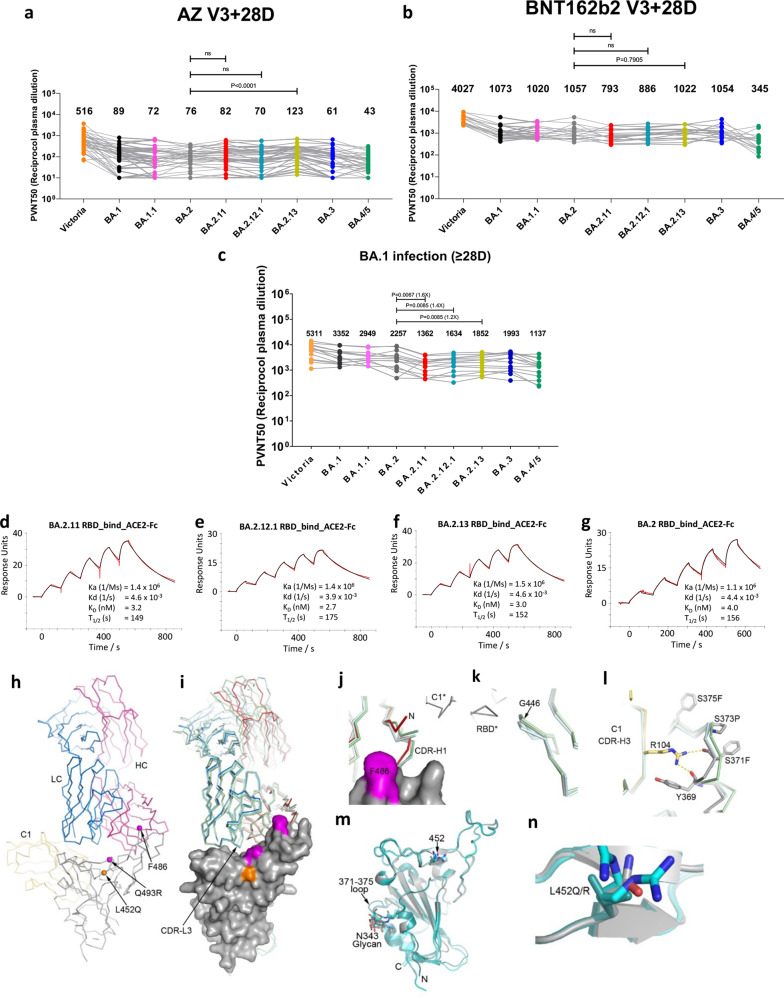
Fig. 1. Characterization of BA.2.11, BA.2.12.1 and BA.2.13 by pseudoviral neutralization assays, SPR, and structural analysis.
a, b IC50 values for the indicated viruses using serum obtained 4 weeks after a third dose of vaccine (a) AstraZeneca AZD1222 (n = 41), b Pfizer BNT162b2 (n = 18). c Neutralization titers of serum from vaccinated volunteers suffering breakthrough BA.1 infection. Comparison is made with neutralization titers to Victoria (SARS-CoV-2/human/AUS/VIC01/2020), BA.1, BA.1.1, BA.2, BA.3, and BA.4/5 as previously reported3. Geometric mean titers are shown above each column. The Wilcoxon matched-pairs signed-rank test was used, and two-tailed P values calculated. d–g SPR sensorgrams (red: experimental binding curve; black: fitted curve) showing ACE2 binding of RBD of BA.2.11 (d), BA.2.12.1 (e), BA.2.13 (f) in comparison with binding to BA.2 RBD (g) (kinetics data shown). Data for BA.2 RBD were reported previously2. h–n Crystal structure of BA.2.12.1 RBD/Beta-27/NbC1 complex. Cα traces with RBD (gray), Beta-27 HC (red) and LC (blue), and NbC1 (yellow). Cαs of L452Q, F486 and Q493R (L, F, and R in BA.2, R, V, and Q in BA.4/5, respectively) shown as spheres (h). Comparison of Beta-27 binding modes between BA.2.12.1 RBD/Beta-27/NbC1 (RBD as surface representation, HC red and LC blue), BA.4/5 RBD/Beta-27/NbC1 (cyan, PDB 7ZXU) and Beta RBD/Beta-27 (green, PDB 7PS1) complexes (RBDs are superposed). Apart from the flexible N- and C-terminal regions of RBD, significant differences occur at the N-terminus and CDR-H1 of the Fab HC, α2 helix, 371–375 loop, and G446 loop of the RBD. CDR-L3 has two conformations in the BA.4/5 RBD complex, and one in other complexes (i). The HC N-terminus and CDR-H1 which contacts RBD residue 486 differs in both Beta and BA.4/5 RBD complexes, the later contains the F486V mutation. Differences are likely caused by contacts from a symmetry-related C1 nanobody shown as gray bonds (j). A structural difference in the G446 loop in the BA.4/5 RBD is induced by crystal contacts (k). The 371–375 loop carrying S371F, S373P, and S375F mutations in BA.2.12.1 and BA.4/5 RBDs is stabilized by interactions with NbC1 CDR-H3 (l). Superimposition of BA.2.12.1 (gray), BA.2 (green, PDB 7ZF9), and BA.4/5 (cyan) RBDs (m). 452 mutations do not introduce significant local structural changes. R452 in BA.4/5 has two conformations (n).