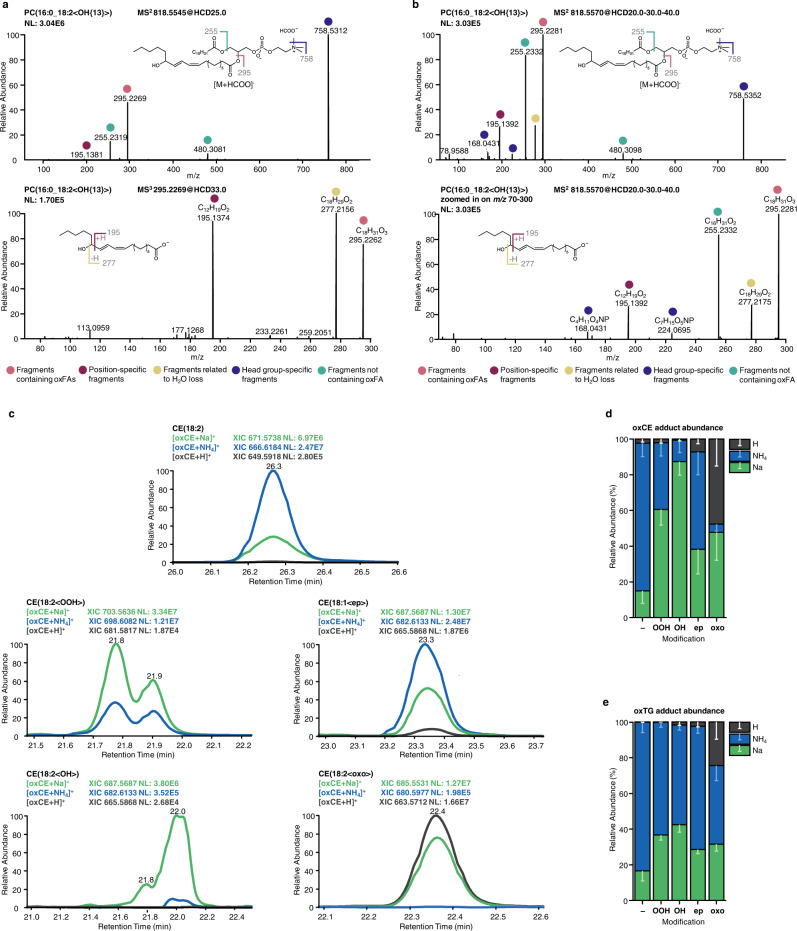
Fig. 1. Optimization of MS and MS2 parameters for the structural elucidation of oxidized complex lipids structures.
Tandem mass spectrometry analysis of oxidized PC lipid using multistage MS3 (a) or optimized elevated energy stepped HCD MS2 experiments (b) both allowed structural assignment of PC(16:0_18:2<OH{13}>). Positive ion mode ionization adducts for in vitro oxidized neutral lipids showed modification type-specific preferences. Qualitative (c; illustrated using extracted ion chromatograms for oxCE(18:2) species) and quantitative (d, e) distributions illustrate modification-specific preferences to form protonated (dark gray), ammoniated (blue) or sodiated (green) adducts for unmodified (-), hydroperoxy- (OOH), hydroxy (OH), epoxy (ep), or keto (oxo) derivatives of CE and TG. Relative abundances of differential adducts for oxCE (d) and oxTG (e) represent (mean ± SD) values calculated for 10 species. Source data are provided as a Source Data file.