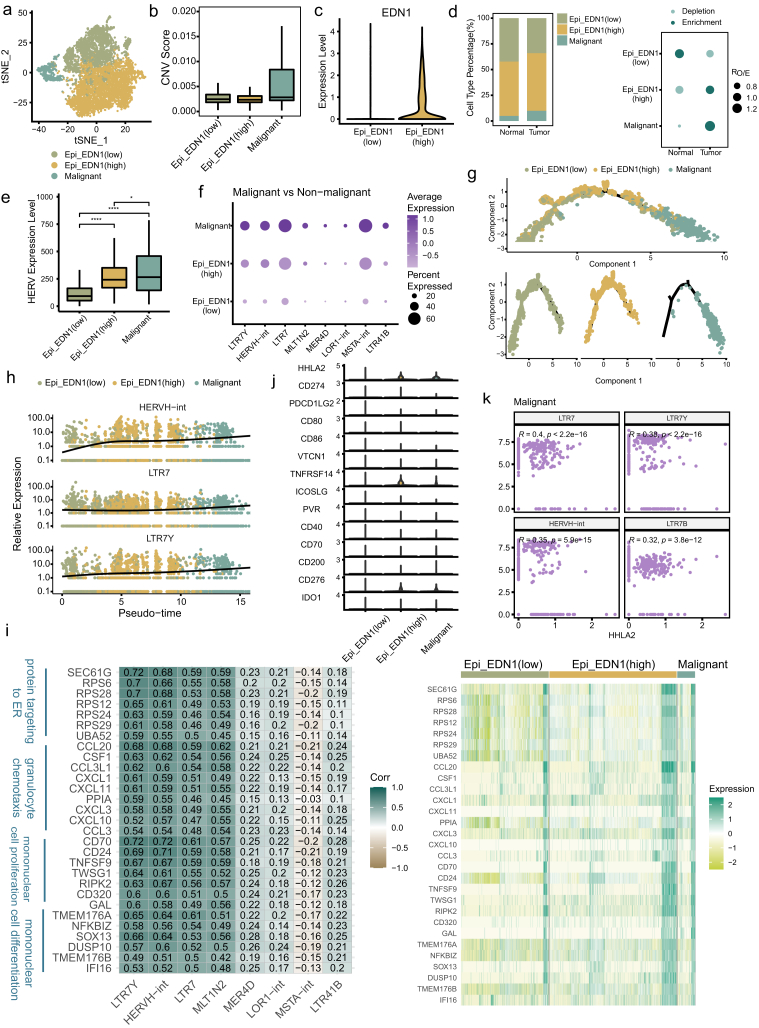
Fig. 3.
Identification of malignant cells and HERVH derepression in the process of malignant transformation. a tSNE projection of epithelial cells, color-coded by cell type. b Boxplot showing the CNV score for each epithelial cell subtype. c Violin plot displaying expression of EDN1 in two non-malignant subtypes. d Relative proportion of epithelial cell subtypes in different tissue types (left). Dot plot showing the distribution of epithelial cell subtypes in different tissue types estimated by Ro/e (right). e Comparison of overall expression of HERVs among epithelial cell subtypes. Statistical significance was evaluated by two-sided Wilcoxon rank sum test. f HERV families upregulated in malignant cells compared to non-malignant cells (adjusted p-value < 0.05 and log2FC > 0.5). g Differentiation trajectory of malignant and normal epithelial cells inferred by Monocle2, color-coded by cell type. h Dynamic expression changes of HERVH elements along the differentiation trajectory. i Heatmaps depicting expression correlation between gene and HERV family in malignant cells (left, Spearman correlation coefficient was also shown) and relative expression of these correlating genes across epithelial cell subtypes (right). j Violin plot displaying expression of the collected immune checkpoints in epithelial cell subtypes. k Scatter plot showing expression correlation between HHLA2 and HERVH element. Spearman correlation coefficient and two-tailed p-value were shown.