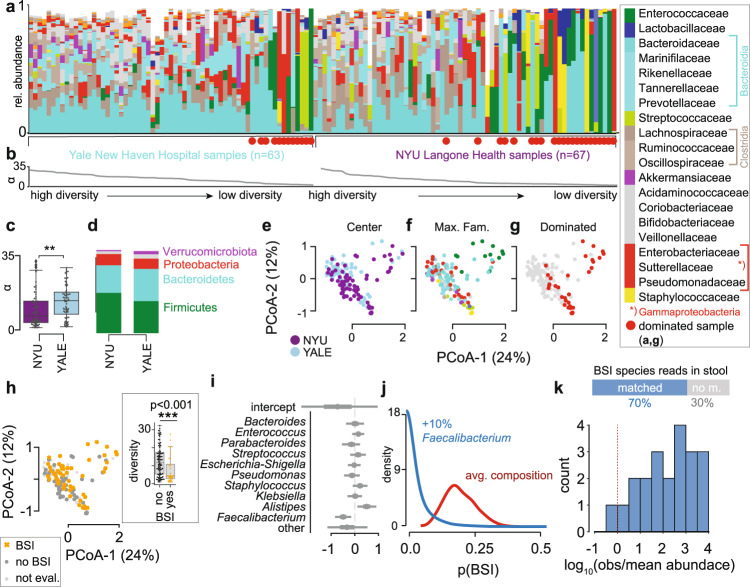
Fig. 3. The dysbiotic gut microbiome in COVID-19 in patients from NYU Langone Health (n = 60) and Yale New Haven Hospital (n = 36) is associated with secondary bloodstream infections.
a Bacterial family composition in stool samples (Yale, n = 63 samples; NYU, n = 67) identified by 16S rRNA gene sequencing; bars represent the relative abundances of bacterial families; red circles indicate samples with single taxa >50%. Samples are sorted by center and bacterial α-diversity (inverse Simpson index, b). c α-diversity in samples from NYU Langone Health and Yale New Haven Hospital; p = 0.0065, two-sided T-test. d Average phylum level composition per center. Principal coordinate plots of all samples shown in a, labeled by center (e), most abundant bacterial family (f) and domination status of the sample (g), and BSI status; inset: boxplot of inverse Simpson index diversity by BSI (h). i Coefficients from a Bayesian logistic regression with most abundant bacterial genera as predictors of BSI status (circle: posterior mean, lines: 95% HDI). j Counterfactual posterior predictions of BSI risk based on bacterial composition contrasting the predicted risk of the average composition across all samples (red) with the risk predicted from a composition where Faecalibacterium was increased by 10% (blue). k Shotgun metagenomic reads matched the species identified in clinical blood cultures in 70% of all investigated cases; the histogram shows the distribution of log10-ratios of relative abundances of matched species in corresponding stool samples to their corresponding mean abundances across all samples.