Abstract
In children undergoing genetic testing for physical health concerns, we examined how often the results also revealed information about their risk for neurodevelopmental disorders. The study sample consisted of 3056 genetic tests (1686 chromosomal microarrays––CMAs, and 1378 next-generation sequencing––NGS panels) ordered at a tertiary pediatric hospital because of a physical/congenital health problem. Tests ordered to investigate developmental concerns were excluded. Pathogenic, or likely pathogenic variants were manually reviewed for diagnostic likelihood, and for evidence of an association with a neurodevelopmental disorder (e.g., autism or intellectual disability). A total of 169 CMAs (10%) and 232 NGS panels (17%) had likely diagnostic results. More than half (52%) of all diagnostic results had established evidence of a neurodevelopmental disorder association. In summary, there is a high prevalence of neurodevelopmental implications from genetic tests ordered for physical/congenital indications. This broad clinical utility suggests a growing need for genetics-first developmental care pathways.
Subject terms: Genetics research, Neurodevelopmental disorders
Introduction
Over the last decade, genetic testing in the form of chromosomal microarray analysis (CMA) and next-generation sequencing (NGS) panels have become routine diagnostic investigations for many health concerns in children. At the same time, our understanding of the genetic architecture of brain development and neurodevelopmental disorders (including autism and intellectual disability) has vastly advanced, with over 1000 associated genes identified.
It has long been understood that many genes impact the development and function of multiple organ systems (e.g., brain and heart) simultaneously, a phenomenon known as pleiotropy. As evidence to this, gene expression analysis has shown that approximately 85% of human protein coding genes are expressed in the brain at some point during development [1, 2], and epidemiologic data consistently support an association between congenital anomalies and neurodevelopmental disorders [3–6].
Consequently, genetic testing ordered to investigate a specific physical or congenital health concern may have significant and potentially unanticipated implications about a child’s future neurodevelopmental trajectory. For example, for infants born with a congenital cardiac malformation, neonatal hypotonia, or a cleft lip/palate, genetic investigations routinely identify variants associated with increased rates of autism, intellectual disability, or adult-onset psychotic disorders [7, 8]. Despite the seemingly obvious nature of this association, the prevalence of this phenomenon has not been estimated. The objective of this study was to determine in what proportion of children undergoing genetic testing for a physical/congenital indication is a genetic variant identified that reveals information about their neurodevelopment.
Methods
Study population
A retrospective review was completed using clinical databases from the molecular genetics laboratory at a tertiary pediatric hospital. The study was approved by the institution Research Ethics Board. The study population included children (≤18 years) who underwent CMA between 2011 and 2017 (inclusive) and/or NGS panel testing between 2015 and 2018 (inclusive) that was ordered by a physician at the same site.
Reason for testing
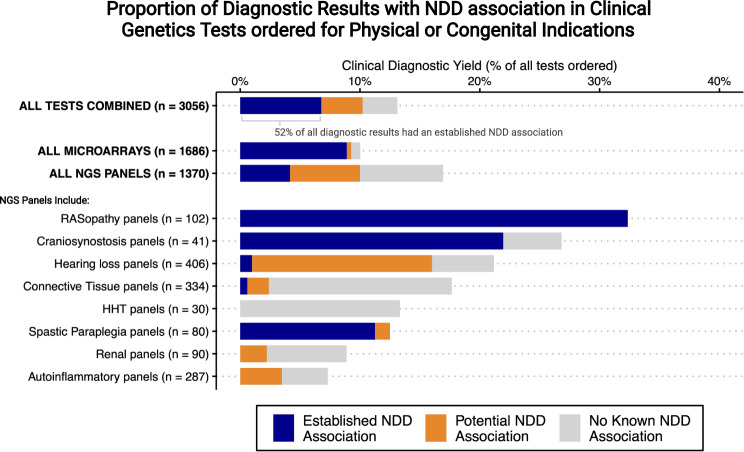
We restricted the sample to genetic tests ordered for a physical or congenital indication (as opposed to a developmental indication). For CMAs, based on provincial funding protocols, we restricted to tests where the ordering physician selected ‘two or more congenital anomalies’ as the reason for testing. Details on the specific congenital anomaly type were not available. We excluded CMAs ordered for the other two possible indications: ‘developmental delay or intellectual disability’ or ‘developmental delay/intellectual disability AND additional clinical features.’ For NGS panel testing, we used a convenience sample including all internally processed genome diagnostic tests at our center (n = 45 total) (see Supplemental methods). We then excluded: (1) single gene tests (e.g., cystic fibrosis, CFTR sequence analysis) (2), panels for syndromes/presentations characteristically associated with neurologic phenotypes (e.g., Batten’s disease) and (3) panels with data sparsity (e.g., ≤5 tests ordered during the study window, n = 2). This yielded a total of eight NGS panels: of these, there were seven where the primary indication for testing was likely to be a physical as opposed to developmental concern (Fig. 1) and one panel (RASopathy) where the indication for testing was possibly physical or developmental (Fig. 1).
Fig. 1. Bar totals show diagnostic yield of individual tests; bars are subdivided by colour to show the proportions with a neurodevelopmental disorder (NDD) association among those with positive results (see Box 1 for criteria).
Full data on counts and panels are available in supplemental materials. Figure created with R and with BioRender.com. Abbreviations: Microarray: chromosomal microarray analysis; NDD: neurodevelopmental disorder; NGS: next generation sequencing panel; HHT: hereditary hemorrhagic telangiectasia; Renal panels: focal segmental glomerulosclerosis/ atypical hemolytic uremic syndrome / membranoproliferative glomerulonephritis.
Variant calls
Variants considered pathogenic or likely pathogenic based on American College of Medical Genetics and Genomics guidelines [9, 10], were manually adjudicated by genetic professionals (genetic counselor, laboratory director, and medical geneticist). First, clinical diagnostic likelihood was determined by considering the requisition indication, the mode of inheritance as specified in OMIM® (omim.org), focused literature reviews, and manual review by the laboratory geneticist and clinical geneticist. Single heterozygous variants in genes associated with autosomal recessive disorders were considered non-diagnostic. Next, likely diagnostic variants were then categorized as having ‘established,’ ‘potential’ or ‘no known’ evidence of a neurodevelopmental disorder association, through review of the published literature and genetic databases, including published case reports or/and clinical records (e.g., listed in ClinVar). Specific criteria for categorization of a neurodevelopmental disorder association are described in Box 1.
Box 1 Criteria for categorization of a neurodevelopmental disorder association (established or potential association) by test type (CMA or NGS panel). CMA chromosomal microarray analysis; NGS next generation sequencing panel.
| Test | Evidence of established neurodevelopmental association | Evidence of potential neurodevelopmental disorder association |
| CMA |
Variant associated with 1) a known syndrome with a neurodevelopmental disorder phenotype, and/or 2) published case reports or clinical records (e.g., listed in ClinVar) of the variant in individuals who have been diagnosed with a neurodevelopmental disorder |
1) Autosomal deletion greater than 3.0 Mb or duplication greater than 10.0 Mb, but similar variants have not been characterised in the medical literature. |
| NGS panels |
1) Gene has been included on 2 or more commercial laboratory panels advertised for autism/ intellectual disability, and/or 2) Gene encodes a protein that is known to play a role in central nervous system development/ function, and/or 3) Variant associated with a clinical syndrome not classically associated with neurodevelopmental disorders but where case reports of such have been described. |
Analysis
Frequencies and proportions were used to summarize findings; a binomial calculation with a normal approximation was used to estimate the 95% confidence intervals. Given that our sample was selected based on indication for testing which would not rule-out a concurrent already identified neurodevelopmental disorder, we also conducted a sensitivity analysis restricting to children under the age of 1 year, where a pre-existing developmental diagnosis would be less likely.
Results
The results from 1686 CMAs included 169 (10%) likely diagnostic variants (Fig. 1; Table S1). Of these, 150 (89%; 95% CI, 83–93%) had established evidence of a neurodevelopmental disorder association. Results were similar after restricting to the 1054 tests performed for children ≤1 year of age (11% diagnostic yield, of which 89% had an established neurodevelopmental disorder association, Table S1).
The results from 1370 NGS panel tests included 232 (17%) likely diagnostic variants, of which 57 (25%; 95% CI, 19–30%) had established evidence of a neurodevelopmental disorder association. Including variants with potential evidence of a neurodevelopmental disorder association increased this proportion to 59% (95% CI, 52–65%) (Fig. 1). Given the large contribution of the RASopathy panel to the proportion with a neurodevelopmental disorder association and given that the indication for ordering this panel was not clearly tied to either a physical or developmental concern, we also estimated overall results excluding this panel. We found only slightly lower proportions (16% diagnostic yield, of which 18% had an established neurodevelopmental disorder association) (Table S1). Likewise, restricting the NGS-panels to the 165 tests performed for children ≤1 year of age yielded slightly higher estimates (29% diagnostic yield, 40% with an established neurodevelopmental disorder association).
Overall, 9% (95% CI, 8–10%) of all CMA tests ordered, 4% (95% CI, 3–5%) of all NGS panel tests ordered, and more than half (52%) of all likely diagnostic results had established evidence of a neurodevelopmental disorder association (Fig. 1 and Supplemental Excel file). Selected examples are discussed in Table 1.
Table 1.
Case Examples of Genetic Testing for a Physical or Congenital Concern Yielding Variants with Neurodevelopmental Implications.
| Clinical example | Test ordered | Result & diagnosis | Developmental implication |
|---|---|---|---|
| Neonate with macrocephaly and polydactyly | CMA | 16p11.2 microdeletion (MIM #611913) | Average IQ 70–80, language delay in ~70%, autism in ~20% |
| Infant with congenital heart defect and cleft palate | CMA | 22q11.2 deletion (MIM #188400) | Average IQ 70–80, autism in ~20%, schizophrenia in ~25% of adults |
| 4-year-old with insidious leg spasticity | Hereditary spastic paraplegia panel | Pathogenic variant in SPG4 (Spastic paraplegia 4) (MIM #182601) | Associated with executive function deficits, behavioural and learning difficulties, intellectual disability in ~4% |
| Infant with feeding difficulties and distinct facial features | RASopathy panel | Pathogenic variant in PTPN11 (Noonan syndrome) (MIM #176876) | Learning disability in ~25%, language and articulation difficulties more common, possibly increased rates of social difficulties and anxiety |
| Infant with craniosynostosis | Craniosynostosis panel | Pathogenic variant in FGFR3 (Muenke syndrome) (MIM #602849) | intellectual disability in ~40%, developmental and/or speech delay in ~65%, ADHD in ~25% |
Note: Clinical examples are for illustrative purposes and have been modified to protect anonymity. See Online Mendelian Inheritance in Man (OMIM®) omim.org for clinical descriptions and literature review for each gene.
ASD Autism Spectrum Disorder, CMA chromosomal microarray analysis, IQ intelligence quotient.
Discussion
Results from over 3,000 routine genetic tests ordered for diagnostic clarification of a physical/congenital concern had clinically relevant neurodevelopmental implications for 5 to 10% of all children tested, and for over half of those with diagnostic results.
Given the increasing availability and resolution of genetic testing across various clinical settings [11], and the recent shift towards genome wide sequencing in very early life [12, 13], the true scope of this phenomenon may be even larger and is likely increasing. Advocates of routine genome sequencing in neonates, both at a general population level [14, 15], or in specific clinical settings [13], highlight potential cost savings [16] and reduced morbidity for those affected with rare and treatable genetic conditions. Our findings emphasize an added level of complexity with respect to neurodevelopmental prognostication. This novel scenario requires additional consideration and research into ethical, psychosocial, and health system implications.
The importance of avoiding delays in access to early interventions and developmental supports in neurodevelopmental disorders is well recognized [17, 18]. Our preliminary results highlight an expanding opportunity for a genetics-first and longitudinal approach to neurodevelopmental research and clinical care. Healthcare institutions, early intervention programs and pre-symptomatic screening and intervention studies may consider formally expanding developmental and mental health care pathways to include those with identified neurodevelopmental disorder variants. This can help to anticipate care needs analogous to programs for other at-risk groups [e.g., infants born prematurely [19], or siblings of those with neurodevelopmental disorders [20]]. In genetics research, this approach can also help address ascertainment bias with respect to neurodevelopmental phenotypes.
Strengths of this study include the large real-world clinical sample, and the consistency of findings across sensitivity analyses. Limitations include reliance on physician order requisitions to categorize test indication and neurodevelopmental history, and the combined estimates across heterogeneous conditions/ tests. Further research is needed to confirm results using genome wide technologies in clinically characterized cohorts, and to further quantify the potential clinical utility of an early genetic diagnosis with respect to anticipatory developmental care.
The overall implication of this study is that the genomic era in neurodevelopmental disorders is likely unavoidable, due to an emerging cohort of children undergoing genetic testing for physical/congenital indications who will be identified somewhat incidentally as being at elevated risk for neurodevelopmental disorders, potentially very early in life. Despite the intuitive nature of this association, its frequency (approximately half of all diagnostic results) has not been previously quantified. Findings support the growing potential of, and need for, genetics-first developmental care pathways in early childhood.
Supplementary information
Acknowledgements
DB acknowledges the Canadian Institutes of Health Research graduate scholarship and the O’Brien scholars program for supporting her fellowship training. SWS holds the Northbridge Chair in Paediatric Research at the Hospital for Sick Children. JV acknowledges the support from the Canadian Institutes of Health Research (PJT-162323).
Author contributions
DAB and JV and NH conceptualized and designed the study, drafted the initial protocol and manuscript, and revised the manuscript. DJS coordinated and supervised data collection and extraction, supervised the genetic analyses/ variant calls, and critically reviewed the manuscript. GC, DAB and NH conducted the genetic analyses/variant calls and critically reviewed the manuscript. DAB, NH and TS extracted and analyzed the data. SWS, DAB, NH, and TS critically reviewed the manuscript for important intellectual content and scientific rigour. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Data availability
Data generated and analysed during this study are included in this published article in the supplementary information files; some restrictions have been applied to de-identify variants and protect patient anonymity.
Competing interests
JV serves as a consultant for NoBias Therapeutics Inc. SWS is on the scientific advisory committee of Population Bio and is an academic consultant of the King Abdullaziz University. The other authors have no relevant conflicts of interest.
Ethical approval
The study was approved by the institution Research Ethics Boards.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-022-01181-z.
References
- 1.Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–9. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Negi SK, Guda C. Global gene expression profiling of healthy human brain and its application in studying neurological disorders. Sci Rep. 2017;7:897. doi: 10.1038/s41598-017-00952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigmon ER, Kelleman M, Susi A, Nylund CM, Oster ME. Congenital heart disease and autism: a case-control study. Pediatrics. 2019;144:e20184114. doi: 10.1542/peds.2018-4114. [DOI] [PubMed] [Google Scholar]
- 4.Timonen-Soivio L, Vanhala R, Malm H, Leivonen S, Jokiranta E, Hinkka-Yli-Salomäki S, et al. The association between congenital anomalies and autism spectrum disorders in a Finnish national birth cohort. Dev Med Child Neurol. 2015;57:75–80. doi: 10.1111/dmcn.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tillman KK, Hakelius M, Höijer J, Ramklint M, Ekselius L, Nowinski D, et al. Increased risk for neurodevelopmental disorders in children with orofacial clefts. J Am Acad Child Adolesc Psychiatry. 2018;57:876–83. doi: 10.1016/j.jaac.2018.06.024. [DOI] [PubMed] [Google Scholar]
- 6.Berg E, Haaland OA, Feragen KB, Filip C, Vindenes HA, Moster D, et al. Health status among adults born with an oral cleft in Norway. JAMA pediatrics. 2016;170:1063–70. doi: 10.1001/jamapediatrics.2016.1925. [DOI] [PubMed] [Google Scholar]
- 7.McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, et al. 22q11.2 deletion syndrome. Nat Rev Dis Prim. 2015;1:15071. doi: 10.1038/nrdp.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler MG, Miller JL, Forster JL. Prader-Willi syndrome-clinical genetics, diagnosis and treatment approaches: an update. Curr Pediatr Rev. 2019;15:207–44. doi: 10.2174/1573396315666190716120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–23. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–5. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 11.Manickam K, McClain MR, Demmer LA, Biswas S, Kearney HM, Malinowski J, et al. Exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability: an evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet in Med. 2021;2029–37. [DOI] [PubMed]
- 12.Dimmock DP, Clark MM, Gaughran M, Cakici JA, Caylor SA, Clarke C, et al. An RCT of rapid genomic sequencing among seriously ill infants results in high clinical utility, changes in management, and low perceived harm. Am J Hum Genet. 2020;107:942–52. doi: 10.1016/j.ajhg.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrikin JE, Cakici JA, Clark MM, Willig LK, Sweeney NM, Farrow EG, et al. The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom Med. 2018;3:6. doi: 10.1038/s41525-018-0045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm IA, Agrawal PB, Ceyhan-Birsoy O, Christensen KD, Fayer S, Frankel LA, et al. The BabySeq project: implementing genomic sequencing in newborns. BMC Pediatr. 2018;18:225. doi: 10.1186/s12887-018-1200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biesecker LG, Green ED, Manolio T, Solomon BD, Curtis D. Should all babies have their genome sequenced at birth? Bmj. 2021;375:n2679. doi: 10.1136/bmj.n2679. [DOI] [PubMed] [Google Scholar]
- 16.Bennette CS, Gallego CJ, Burke W, Jarvik GP, Veenstra DL. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2015;17:587–95. doi: 10.1038/gim.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller EA, Kaiser AP. The effects of early intervention on social communication outcomes for children with autism spectrum disorder: a meta-analysis. J autism developmental Disord. 2020;50:1683–700. doi: 10.1007/s10803-019-03927-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McManus BM, Richardson Z, Schenkman M, Murphy N, Morrato EH. Timing and intensity of early intervention service use and outcomes among a safety-net population of children. JAMA Netw Open. 2019;2:e187529–e. doi: 10.1001/jamanetworkopen.2018.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soleimani F, Azari N, Ghiasvand H, Shahrokhi A, Rahmani N, Fatollahierad S. Do NICU developmental care improve cognitive and motor outcomes for preterm infants? A systematic review and meta-analysis. BMC Pediatrics. 2020;20:67. doi: 10.1186/s12887-020-1953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JZ, Iverson JM, Roemer EJ, Plate S, Schneider JL. “I’m Worried About My Child”: a longitudinal investigation of parental concerns and repeat screening in toddlers with familial risk of autism spectrum disorder. Pediatrics. 2021;147:73–4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated and analysed during this study are included in this published article in the supplementary information files; some restrictions have been applied to de-identify variants and protect patient anonymity.