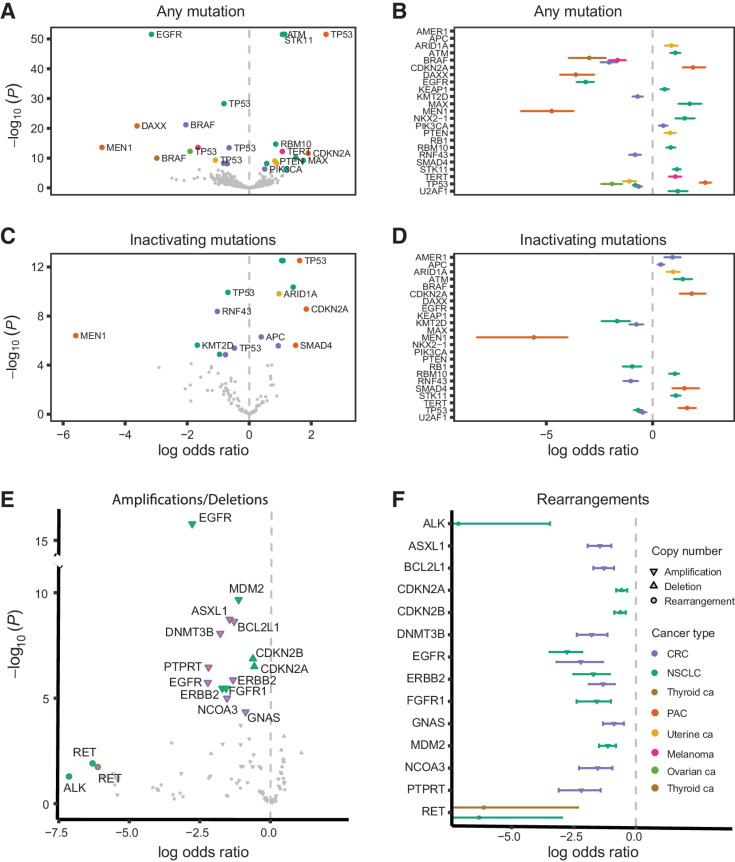
Figure 3.
Co-occurrence of RAS hotspot mutations with sequence and structural non-RAS mutations is cancer lineage specific. A, Volcano plots of the posterior median log OR (x-axis) versus negative log10P value (y-axis) for the association between RAS mutations and non-RAS variants. Mutations in non-RAS genes with log OR greater than 0 co-occur with RAS mutations as opposed to non-RAS mutations with an interaction coefficient less than 0, which are comutated with RAS at a rate lower than expected or are mutually exclusive. The further to the right on the x axis the closest to true co-occurrence and the further to the left of the axis the closest to true mutual exclusivity, with the statistical significance of the difference from 0 (which indicates independence) plotted on the y-axis. B, For statistically significant associations, 95% posterior credible intervals of the log OR are indicated by error bars. For genes associated with RAS mutations in multiple cancers, multiple vertically offset error bars are displayed. C and D, Volcano plots and posterior credible intervals for the association between RAS mutations and inactivating non-RAS mutations. E, Volcano plots of the posterior median log OR (x-axis) versus negative log10P (y-axis) for the association between RAS mutations with deep deletions, high copy amplifications, and gene fusions. F, 95% posterior credible intervals of the log OR for RAS/non-RAS fusions. Despite the small number of fusions included in the analyses that precluded firm statistical conclusions, fusions involving ALK (EML4-ALK) and RET (RET-KIF5B) genes showed a pattern of mutual exclusivity with RAS mutations. CRC, colorectal cancer.