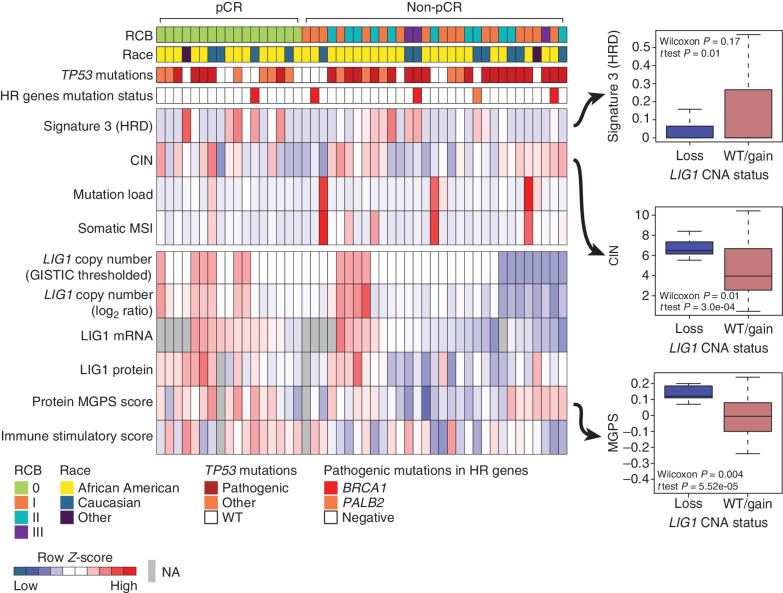
Figure 5.
Proteogenomic features associated with LIG1. Heat map showing copy number, mRNA, and protein levels of LIG1, which are significantly (Wilcoxon rank sum test) lower (blue) in non-pCR tumors. Corresponding box plots show that tumors with low-level copy loss of LIG1 (GISTIC = −1, likely single copy-number loss) display significantly higher chromosomal instability and MGPS and significantly lower signature 3 (COSMIC mutational signature associated with HRD) than tumors that are wild-type (WT) or show gain of CNA (GISTIC ≥0). Wilcoxon rank sum tests and t tests were used to compare LIG1-loss cases with LIG1-intact (WT/gain) cases. HR, homologous recombination; MSI, microsatellite instability; NA, not available.