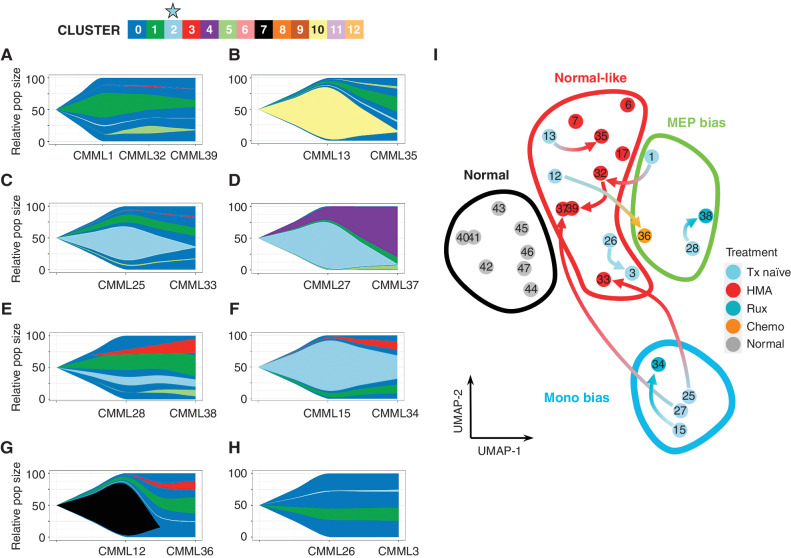
Figure 5.
Hypomethylating agent treatment is associated with the depletion of Clus2 cells. Although mitoClone revealed stable clonality distribution after treatment, as demonstrated in Fig. 4L–S, the cluster dynamics changed dramatically after treatment with (A–D) HMA, (E–F) ruxolitinib, (G) chemotherapy, and (H) no treatment. Interestingly, two HMA-treated patients (C–D) were monocytic-biased in their pretreatment sample and showed a marked decrease in cluster 2 fraction (light blue) following HMA treatment. I, Sample movement in pseudo-bulk representation on UMAPs following treatment (complementary analysis in Supplementary Fig. S2; sample numbering matches the sample numbering in Fig. 4L–S and Fig. 5A–H). Three of the four pre-HMA treatment samples move (red arrows) from monocytic-biased or MEP-biased to normal-like phenotype after treatment with HMA. Additionally, three samples were collected from patients after HMA treatment (without a matched treatment-naïve sample) that were also grouped in this normal-like phenotype. Ruxolitinib treatment shows some change in transcriptomics but is not sufficient to be classified with a new differentiation trajectory following treatment.