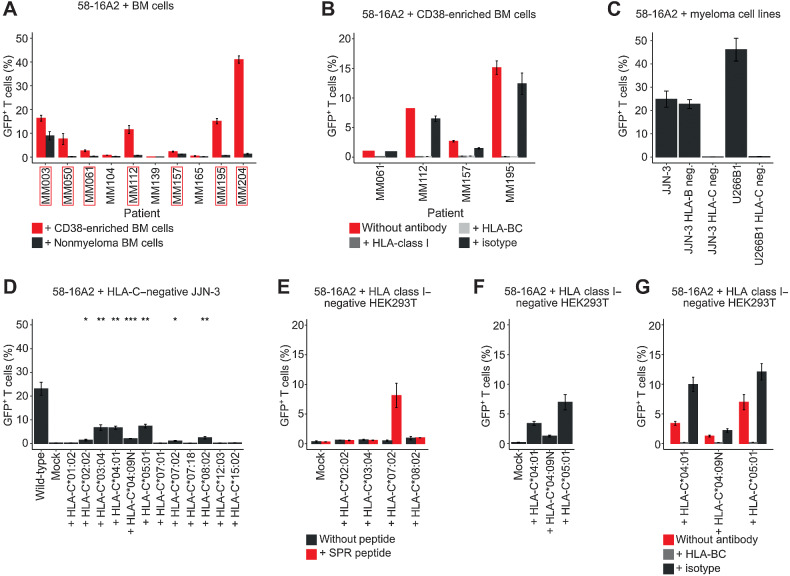
Figure 5.
16A2 is a degenerate TCR that recognizes multiple myeloma cells of different patients in the context of a variety of HLA-C alleles. A, 58-16A2 were incubated with CD38-enriched multiple myeloma or MACS CD38-depleted nonmyeloma cells. Axis descriptions of patients who activated 58-16A2 are highlighted by red rectangles. “Nonmyeloma BM cells” represent CD38-depleted bone marrow (BM). In patients MM157 and MM195, remaining multiple myeloma/B-lineage cells were removed from MACS CD38-depleted bone marrow by additional FACS sorting (sorting criteria: CD38−CD138−CD19−, gating in Supplementary Fig. S8). B, 58-16A2 were incubated for 16 hours at 37°C in 5% CO2 with multiple myeloma cells from four patients in the presence of blocking antibodies against HLA-class I or HLA-BC. C, 58-16A2 were incubated with JJN-3 or U266B1 cells for 16 hours at 37°C in 5% CO2. HLA-B or HLA-C were deleted using CRISPR-Cas9. D, 58-16A2 were incubated for 16 hours at 37°C in 5% CO2 with HLA-C–negative JJN-3 transiently transfected with selected HLA-C alleles. Wild-type, unmanipulated JJN-3 expressing all endogenous HLA alleles; mock, nucleofection of HLA-C–negative JJN-3 without HLA-containing plasmid. Statistical significance between mock and indicated conditions was determined by paired Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. E–G, Selected HLA-C alleles were expressed in HLA-class I–negative HEK293T cells and incubated with 58-16A2 for 16 hours at 37°C in 5% CO2. Mock, transfection without HLA-containing plasmid. E, HLA-recombinant HEK293T were also loaded with SPR. G, Blocking antibodies against HLA-BC or an isotype antibody were added before coincubation. Bar charts indicate mean ± standard deviation. Data are representative of 3 experiments.