Fig. 1.
MreB double protofilaments adopt quasi-stable twist states with similar sets of angles across simulations and nucleotide-binding states.
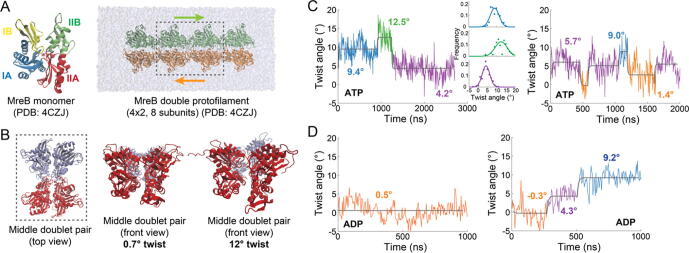
A) Left: Caulobacter crescentus MreB (CcMreB) monomer structure bound to ATP (from filament structure PDB ID: 4CZJ). The four subdomains align with those of actin and are defined by IA (residues 9 to 36 and residues 322 to 345, blue), IB (residues 37 to 81, yellow), IIA (residues 151 to 186 and residues 265 to 321, red), and IIB (residues 187 to 264, green). Right: double protofilament (4x2) structure of CcMreB (PDB ID: 4CZJ), which consists of antiparallel MreB strands (green, orange). The middle doublet (dotted square) was used for twist measurements (Methods). The silver transparent water box is shown to illustrate the size of the simulated system.
B) The twist angle of an MreB double protofilament measures the relative rotation of pairs within the middle doublet. Left: schematic of the middle doublet (2x2) used to calculate the twist angle of MreB double protofilaments. Shown in red and blue are antiparallel subunit pairs. Middle: subunit pairs (red, blue) are shown through the long axis of the double protofilament from the crystal structure (0.7° twist). Right: subunit pairs (red, blue) rotate in opposite directions to generate twist, shown here at 12°.
C) Twist angle varied throughout ATP-bound double protofilament simulations. Left: twist angle throughout a 2.7-ms simulation of an ATP-bound 4x2 MreB structure. Colored segments with overlapping horizontal grey lines represent states identified by the change-point algorithm Steppi (Methods). Inset: the twist angle distribution of each state fit with a Gaussian function. Right: A 2-ms replicate simulation of an ATP-bound 4x2 MreB filament displayed different twist dynamics from the simulation on the left, but the collective set of twist angles was similar in the two simulations. Steppi-identified states with similar mean values between the two simulations are plotted with the same color.
D) Twist angle sometimes varied throughout ADP-bound double protofilament simulations and adopted generally lower values than ATP-bound structures. Left: A 1-ms simulation of an ADP-bound 4x2 MreB double protofilament was characterized by a single state with low twist (orange). Right: the twist angle of a replicate 1-ms simulation of an ADP-bound 4x2 MreB double protofilament varied between three states.