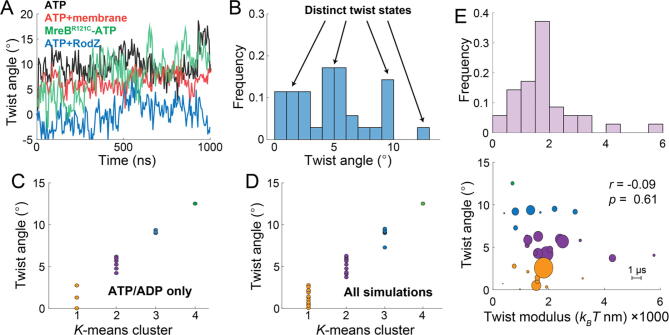
Fig. 2.
MreB double protofilaments occupy at least four distinct twist states and perturbations shift their relative occupancy.
A) Representative 1-ms trajectories of the effects of each perturbation on MreB double protofilament twist behavior. Binding to a membrane (red) decreased twist, while the R121C mutation led to higher twist angles (green). Binding to RodZ (blue) caused the largest reduction in twist angle.
B) The distribution of mean twist angles among Steppi-identified states across all simulations from this study shows multimodal behavior. Modes are identified with arrows.
C) K-means clustering with n=4 of Steppi-identified twist states across ATP- and ADP-bound free (no membrane, no RodZ) double protofilament simulations. Clusters are identified by color and each state instance is shown as a colored circle.
D) K-means clustering with n=4 of Steppi-identified twist states across all simulations. Clusters are identified by color and each state instance is shown as a colored circle.
E) Top: distribution of twist moduli calculated for each state identified by Steppi (Methods). Bottom: twist modulus did not display any obvious trends across twist states. States are colored based on k-means cluster (C,D), and the size of each circle is proportional to the state lifetime (scale shown at bottom right).