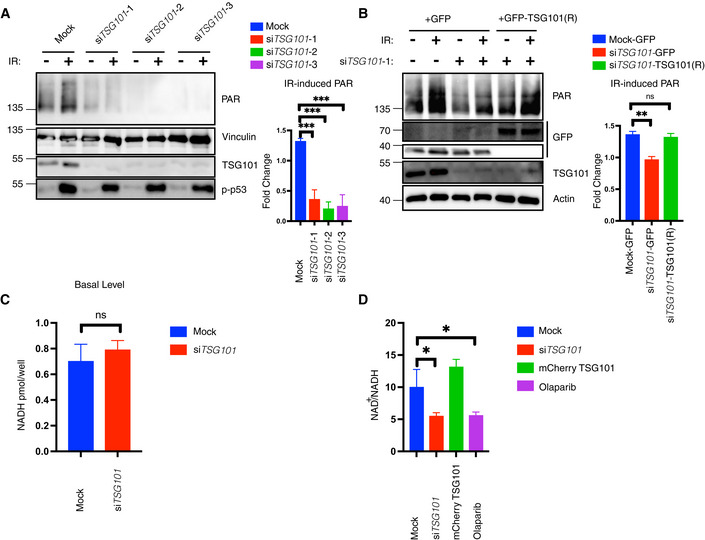
Figure EV3. TSG101 is essential for PARylation.
-
AU2‐OS cells were transfected with nontargeting (mock) or three different TSG101‐targeting siRNAs. Cells were irradiated (20 Gy) 10 min before analysis. Nuclear extracts were immunoblotted with the indicated reagents and antibodies. Right panel, densitometric measurements of irradiation‐induced fold changes in PARylation were obtained from three independent experiments and conditions were compared with an ordinary one‐way ANOVA (***P < 0.001). Error bars represent mean ± SD.
-
BA rescue experiment was performed using a siRNA‐resistant GFP‐TSG101(R) expression vector. Two base substitutions were introduced into the codons for amino acids 61 and 62 of TSG101. U2‐OS cells were transfected with either GFP alone or GFP‐TSG101 (R) vector. Cells were then transfected with nontargeting or TSG101‐targeting siRNAs as indicated and were irradiated (20 Gy) 10 min before analysis. Nuclear extracts were immunoblotted with the indicated reagents and antibodies. Densitometric measurements of irradiation‐induced fold changes in PARylation (right panel) were obtained from three independent experiments. Conditions were compared with an ordinary one‐way ANOVA (ns, P > 0.05; **P < 0.01). Error bars represent mean ± SD.
-
CU2‐OS cells were transfected with nontargeting or TSG101‐targeting siRNAs. NADH measurements were performed 72 h after the siRNA transfection. Conditions were compared with Student's t‐test (Welch Correction) (ns, P > 0.05). Error bars represent mean ± SD. Results were obtained from three biologically independent experiments.
-
DU2‐OS cells were transfected with nontargeting (Mock) or TSG101‐directed siRNAs, with mCherry‐TSG101, or treated with olaparib (10 μM, 16 h before irradiation), as indicated. Cells were irradiated (20 Gy) 10 min before analysis and NAD+/NADH levels were determined. The result is representative of four independent experiments. The conditions were compared with an ordinary one‐way ANOVA (*P < 0.05). Error bars represent mean ± SD.
