Figure 1. A genome‐wide CRISPR screen for SARS‐CoV‐2 entry host factors.
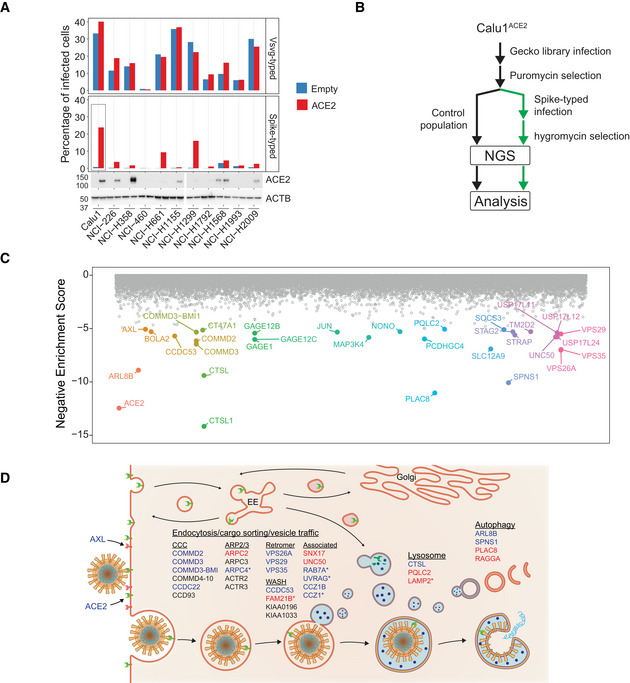
- Infection susceptibility (top) to VSVG and Spike‐typed lentiviruses of a panel of lung cancer cell lines with or without (Empty vector) overexpression of ACE2. ACE2 protein levels were determined by Western blot (bottom).
- Screen strategy: Calu1ACE2 cells were transduced with the GeckoV2 CRISPR library to create a mutant library that was subsequently infected with Spike‐typed lentiviruses carrying a hygromycin‐resistant plasmid. After hygromycin selection, the dropout of sgRNAs in the infected population compared with an uninfected population was assessed by NGS.
- Gene‐level scores obtained by robust ranking algorithm analysis of the sgRNA relative abundances between cells infected with Spike‐typed lentiviruses and uninfected cells. Genes were ordered alphabetically on the x‐axis and the top hits (log10(enrichment‐score) < −5) were colored and labeled. Note that CTSL and CTSL1 are different gene isoforms.
- Screen results integration in pathways previously linked to SARS‐CoV‐2 entry. Black: nonsignificant genes reported by others; Blue: significant genes previously reported; Red: novel genes not reported in previous screens. *Nonsignificant based on FDR (FDR > 0.05) but present in the top 200 hit list.
Source data are available online for this figure.
