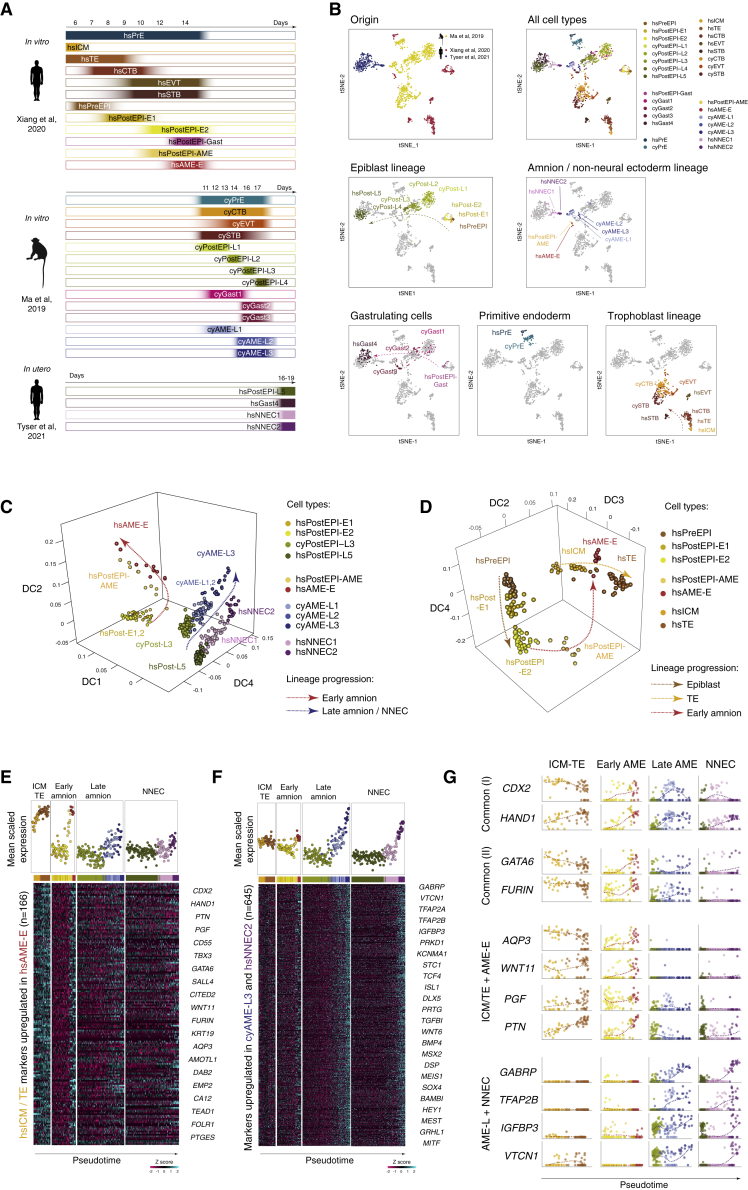
Figure 1.
Two independent trajectories of amniogenesis in primate embryos
(A) Cell types and their developmental stages within the integrated RNA-seq dataset. Note that developmental timing in vitro and in utero may differ.
(B) tSNE of the integrated single-cell RNA-seq dataset of primate embryos.
(C) Diffusion maps of amnion and non-neural ectoderm lineages in primate embryos. (D) Diffusion maps of early amnion and trophectoderm lineages in primate embryo.
(E) Expression of the genes commonly upregulated in hsICM/hsTE and hsAME-E as compared with epiblast in embryo.
(F) Expression of the genes commonly upregulated in cyAME-L3 and hsNNEC2 as compared with epiblast in embryo.
(G) Expression of selected markers during lineages progression in embryo.
See also Figures S1 and S2.