Fig. 2.
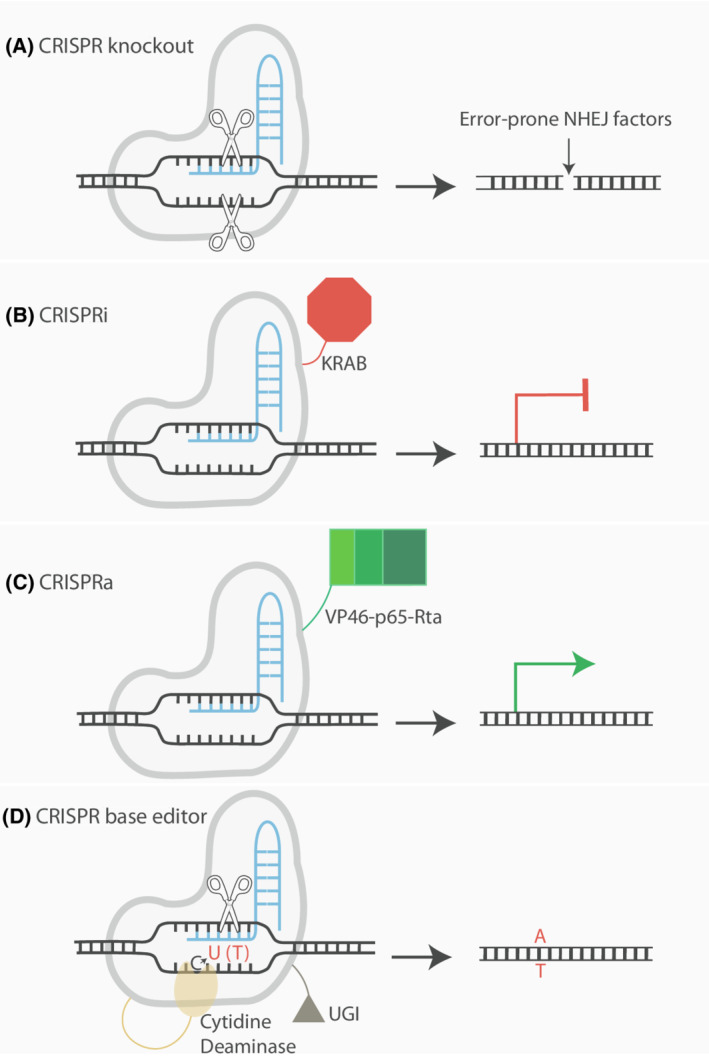
An overview of CRISPR‐Cas9 constructs that are utilized in pooled screens. (A) CRISPR knockout induces a blunt‐ended DNA double‐strand break in a gene of interest. This is then typically repaired by error‐prone non‐homologous end joining (NHEJ) resulting in a frame‐shift mutation. (B) CRISPR interference (CRISPRi) consists of a dCas9 fused with a Krüppel associated box (KRAB) domain which transcriptionally silences the promoter of interest. (C) CRISPR activation (CRISPRa) also consists of a dCas9 which is fused with a VP46 and the activator domains of the transcription factors p65 and Rta. This transcriptionally activates the promoter of interest. (D) Base editor, here presented by a cytidine base editor, allows introduction of single nucleotide variants. The C base is converted into a U base intermediate. The fused uracil DNA glycosylase inhibitor (UGI) domain protects the newly formed uracil intermediate from uracil‐DNA glycosylase. The nickase Cas9 (nCas9) generates a nick on the sgRNA target strand thereby activating mismatch repair (MMR) to excise the nicked target strand base. This converts the original U:G mismatch into a U:A pair. The U is finally converted to a T base via repair or replication. [Colour figure can be viewed at wileyonlinelibrary.com]
