Abstract
Background
The deeper understanding of the complex hereditary basis of familial hypercholesterolemia (FH) has raised the rationale of genetic testing, which has been underutilized in clinical practice.
Objectives
The present study aimed to explore the variant spectrum of FH in an expanding manner and compare its diagnostic performance.
Methods
A total of 169 Chinese individuals (124 index cases and 45 relatives) with clinical definite/probable FH were consecutively enrolled. Next-generation sequencing was performed for genetic analysis of 9 genes associated with hypercholesterolemia (major genes: LDLR, APOB, and PCSK9; minor genes: LDLRAP1, LIPA, STAP1, APOE, ABCG5, and ABCG8) including the evaluations of small-scale variants and large-scale copy number variants (CNVs).
Results
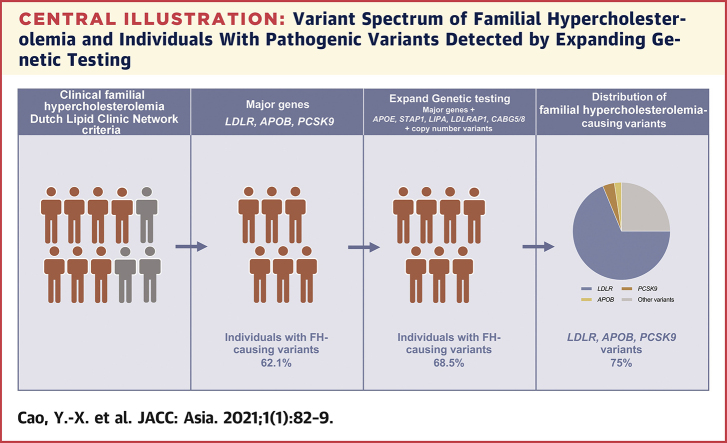
Among the 169 clinical FH patients included, 98 (58.0%) were men. A total of 85 (68.5%) index cases carried FH-associated variants. The proportion of FH caused by small-scale variants in LDLR, APOB, and PCSK9 genes was 62.1% and then increased by 6.5% when other genes and CNVs were further included. Furthermore, the variants in LDLR, APOB, and PCSK9 genes occupied 75% of all FH-associated variants. Of note, there were 8 non-LDLR CNVs detected in the present study.
Conclusions
LDLR, APOB, and PCSK9 genes should be tested in the initial genetic screening, although variants in minor genes also could explain phenotypic FH, suggesting that an expanding genetic testing may be considered to further explain phenotypic FH.
Key Words: copy number variant, familial hypercholesterolemia, genetic testing, variant
Abbreviations and Acronyms: APOB, apolipoprotein B; CNV, copy number variant; CVD, cardiovascular disease; FH, familial hypercholesterolemia; LDL-C, low-density lipoprotein-cholesterol; LDLR, low-density lipoprotein receptor; PCSK9, proprotein convertase subtilisin/kexin type 9
Central Illustration
As one of the most prevalent autosomal codominant inherited disorders, familial hypercholesterolemia (FH) is characterized by extreme low-density lipoprotein-cholesterol (LDL-C) elevation and a high lifetime risk of cardiovascular disease (CVD) (1). According to the recent evidence, the prevalence of heterozygous familial hypercholesterolemia (HeFH) has been estimated to be as high as 1:200 in the general population (2). Despite a relatively low prevalence ranging from 1:160,000 to 1:300,000, the homozygous form exhibits extreme hazard with high CVD morbidity during childhood and premature mortality before age 30 years (3). Thus, timely and accurate diagnosis of FH is of vital importance for the treatment and prevention of severe consequences. Until now, the Dutch Lipid Clinic Network (DLCN) criteria, Simon Broome system, Make Early Diagnosis-Prevent Early Death, and other national diagnostic algorithms have been adopted (1,4). The disunity of clinical diagnosis accentuates the role of genetic screening, which, however, has been underutilized for facilitating a definite diagnosis of FH (5).
As far as we know, FH is mainly caused by the mutation of low-density lipoprotein receptor (LDLR) gene, apolipoprotein B (APOB) gene, or proprotein convertase subtilisin/kexin type 9 (PCSK9) gene in an autosomal dominant pattern. Rarely, FH is caused autosomal recessive mutations in the low-density lipoprotein receptor adaptor protein 1 (LDLRAP1) gene (1). Among them, the LDLR gene has acquired comprehensive investigations, with an identification of more than 2,900 variants in the Leiden Open Variation Database (6). With the advancement of sequencing technique, the knowledge about the molecular basis of FH has been expanded in recent years. To date, more genes, including signal transducing adaptive family member 1 (STAP1), apolipoprotein E (APOE), and lipase A (LIPA), have been identified as possible FH-causing genes (7,8). Furthermore, sitosterolemia, a rare autosomal recessive condition caused by variants in the ATP-binding cassette transporter family members 5 and 8 (ABCG5/8), has similar phenocopies (9). Additionally, the next-generation sequencing (NGS) made it possible to detect copy number variants (CNVs) rapidly and conveniently (10). But, in parallel, the clinical significance of these newly discovered variants is usually uncertain, and their value for genetic diagnosis may be limited, which reinforces the need of further family evaluation and functional studies.
Considering the ethnic heterogeneity of the molecular basis of FH, previous studies from various countries and regions have explored the variant spectrum of FH mainly with the investigations of variants in the LDLR, APOB, and PCSK9 genes (11). Also, in our prior study, mutational analysis revealed a detection rate of 46.9% using target exome sequencing for these 3 genes in China (12). Given that the variants were not detected in nearly one-half of patients, it is imperative to continue to sort out the comprehensive genetic basis of FH in Chinese population. The aim of the current study is to further explore the variant spectrum of FH using the NGS approach in an expanding manner.
Methods
Study design and populations
In the current study, we consecutively recruited 124 index cases and 45 relatives with clinical FH from April 2016 to March 2017 in Fuwai Hospital. After exclusion of secondary hypercholesterolemia caused by severe thyroid, liver, and renal dysfunction, patients were diagnosed as definite or probable FH according to DLCN criteria with a score ≥6 by experienced physicians. Besides, children were considered to be FH if they had values of LDL-C above the 95th percentile according to age and sex and family history of high cholesterol and/or premature familial cardiovascular disease.
Clinical data from each participant were collected by physicians and experienced nurses including family and personal history of dyslipidemia and coronary artery disease (CAD). Family history of CAD was defined as first-degree relatives with known CAD. For precise clinical diagnosis, we have well-documented the untreated and maximum LDL-C levels for each patient at full steam. Nevertheless, some patients reported using lipid-lowering therapy at the time of recruitment without their untreated level of LDL-C. Thus, we had to take into account the effect of lipid-lowering therapy, and adjusted the level of LDL-C according to the dose and potency of statins (13). Furthermore, we uniformly recorded the physical signs including xanthoma and corneal arcus.
Our study complied with the Declaration of Helsinki and was approved by the hospital’s ethical review board (Fu Wai Hospital & National Center for Cardiovascular Diseases, Beijing, China). Informed written consent was obtained from all participants.
Laboratory examination
Blood samples were collected from the cubital vein into ethylenediaminetetraacetic acid–containing tubes for biochemical measurements after a 12-h overnight fast. After centrifugation, plasma total cholesterol, triglycerides, high-density lipoprotein cholesterol, and LDL-C were measured using a standard enzymatic assay with automatic biochemistry analyzer (Hitachi 7150, Tokyo, Japan). Apolipoprotein A (apo A) and apoB were assayed by a commercial available turbidimetric immunoassay.
Genetic sequencing
Briefly, genetic analysis of 9 genes associated with hypercholesterolemia (major genes: LDLR, APOB, and PCSK9; minor genes: LDLRAP1, LIPA, STAP1, APOE, ABCG5, and ABCG8) and their CNVs were performed using NGS for the index cases. Then, their relatives were sequenced in 3 loci or 9 loci according to the results of their index cases.
The blood samples for DNA extraction were well preserved at −80°C after centrifugation for 10 min at 3,500 rpm, 4°C. The genomic DNA was isolated form peripheral blood leukocytes using standard extraction protocols. After quantification of the DNA samples with Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, Delaware), targeted exome Illumina Libraries were prepared according to the manufacturer’s standard protocol (MyGenostics, Beijing, China). Furthermore, the amplified DNA was captured with an FH-related Gene Panel using biotinylated oligo-probes (MyGenostics). The probes were designed to tile along 9 FH-related genes covering the coding exons, intron-exon boundaries, and 3′/5′ untranslated region (UTR) of LDLR, APOB, PCSK9, LDLRAP1, APOE, LIPA, STAP1, and ABCG5/8. Then, the enrichment libraries were sequenced on an Illumina HiSeq 2000 sequencer for paired-end reads of 2 × 100 base pairs.
In silico analysis
After sequencing, high-quality reads were retrieved from raw reads by filtering out the low-quality reads and adaptor sequences using the Solexa QA package and the Cutadapt program (14), respectively. The clean read sequences were then aligned to the human genome reference sequence (hg19) using SOAP aligner program. After the PCR duplicates were removed by the Picard software, the single nucleotide polymorphisms (SNPs) were firstly identified using the SOAP SNP program. Subsequently, we realigned the reads to the reference genome using BWA and identified the insertions or deletions (InDels) using the GATK program. The identified SNPs and InDels were annotated using the Exome-assistant program. MagicViewer was used to view the short-read alignment and validate the candidate SNPs and InDels.
Annotated variants were first filtered and defined as uncommon variants according to minor allele frequencies <1% in the general population of 1KG, Exome Aggregation Consortium, Exome Sequencing Project 6500, and Inhouse databases. Afterward, the variants that have not been recorded in the public database, including Human Gene Mutations Database and ClinVar, were defined as “novel” variants. Sanger sequencing was performed to confirm the novel variants. Bioinformatics tools including PolyPhen-2, Sorting Tolerant From Intolerant and MutationTaster, SPIDEX, and InterVar were used to predict the potential pathogenicity of the novel variants. A novel variant was determined to be pathogenic or likely pathogenic if ≥2 silico algorithm gave deleterious prediction. Double heterozygote was defined as 2 variants in 2 different FH-causing genes, and compound heterozygote was defined as different variants in both alleles of the same gene.
The CNVs were filtered based on a ratio and justified for their function change. A ratio threshold value ≤0.7 was used to identify probable deletions, whereas a ratio value ≥1.3 was used for recognition of duplications (15). The detection rate was defined as the proportion of FH-associated variants in participants.
Statistical analysis
All statistical analysis were performed using SPSS version 21.0 (SPSS Inc., Chicago, Illinois). Continuous variables with normal distribution were presented as mean ± SD, and median (25th to 75th percentile) represented continuous variables with non-normal distribution. The comparisons of continuous parameters were evaluated with Student’ s t-test or Mann-Whitney U test. Categorical variables were presented as number (percentage) and analyzed using the chi-square test. A p value <0.05 was defined as a significant difference.
Results
The clinical characteristics of the cohort were shown in Table 1. The current study recruited 124 index cases and 45 relatives. The mean age of the index cases was 45 ± 14 years and there were more men (62.1%). In addition, physical signs including xanthoma and/or corneal arcus were present in 24 patients (14.2%). In total, 71.0% of the index cases had a personal history of CAD and 38.0% of the participants had a family history of CAD. According to the DLCN criteria, 48 (38.7%) phenotypic index cases were diagnosed as definite FH and the residual 61.3% patients were probable FH. However, there were more probable FH in the group of relatives. Besides, carriers with FH-causing variants were significantly younger and presented with a higher prevalence of physical signs, including xanthoma and corneal arcus (both p < 0.05). In addition, they also had a significantly lower level of triglycerides, high-density lipoprotein cholesterol, and apoA (all p < 0.05).
Table 1.
Clinical Characteristics of the Participants
| Index Cases |
p Value | Relatives (n = 45) | |||
|---|---|---|---|---|---|
| Overall (N = 124) | Variant – (n = 39) | Variant + (n = 85) | |||
| Age, yrs | 45 ± 14 | 48 ± 12 | 43 ± 14 | 0.038 | 42 ± 20 |
| Male | 77 (62.1) | 25 (64.1) | 52 (61.2) | 0.755 | 21 (46.7) |
| Xanthoma/corneal arcus | 22 (17.7) | 2 (5.1) | 20 (23.5) | 0.013 | 2 (4.4) |
| Personal history of CAD | 88 (71.0) | 27 (69.2) | 61 (71.8) | 0.773 | 8 (30.8) |
| Family history of CAD | 46 (38.0) | 16 (44.4) | 30 (35.3) | 0.878 | 31 (68.9) |
| Statin at admission | 87 (70.2) | 27 (69.2) | 60 (70.6) | 0.343 | 11 (24.4) |
| Definite FH | 48 (38.7) | 11 (28.2) | 37 (43.5) | 0.104 | 9 (20.9) |
| LDL-C, mmol/l | 5.81 ± 2.41 | 5.61 ± 2.39 | 5.90 ± 2.42 | 0.536 | 6.14 ± 1.55 |
| HDL-C, mmol/l | 1.11 ± 0.35 | 1.23 ± 0.33 | 1.05 ± 0.34 | 0.006 | 1.31 ± 0.33 |
| TC, mmol/l | 7.66 ± 2.81 | 7.81 ± 2.98 | 7.59 ± 2.74 | 0.707 | 8.20 ± 1.92 |
| Apo A, g/l | 1.28 ± 0.34 | 1.44 ± 0.33 | 1.20 ± 0.32 | <0.001 | 1.35 ± 0.26 |
| Apo B, g/l | 1.51 ± 0.51 | 1.52 ± 0.39 | 1.50 ± 0.55 | 0.834 | 1.51 ± 0.38 |
| Triglyceride, mmol/l | 1.40 (1.02–1.94) | 1.81 (1.37–2.58) | 1.25 (0.94–1.66) | <0.001 | 1.30 (1.01–2.03) |
| Lipoprotein (a), mg/dl | 31.65 (16.26–67.28) | 23.84 (8.17–45.93) | 35.74 (17.31–73.33) | 0.106 | 25.00 (10.51–44.34) |
Values are mean ± SD, n (%), or median (25th–75th percentile).
apo = apolipoprotein; CAD = coronary artery disease; FH = familial hypercholesterolemia; HDL-C = high-density lipoprotein-cholesterol; LDL-C = low-density lipoprotein-cholesterol; TC = total cholesterol.
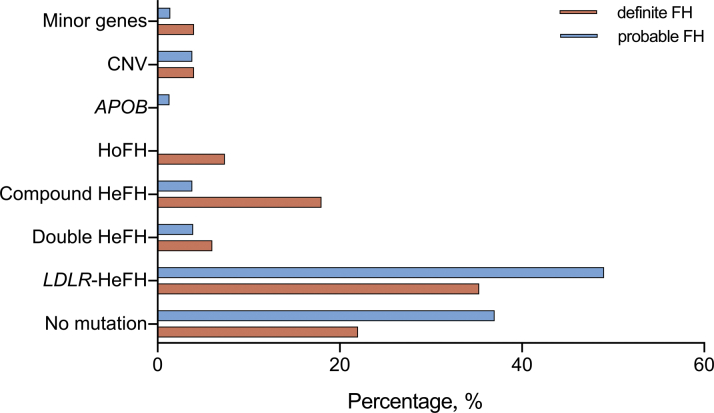
As shown in Table 2, we identified 85 FH-causing variants carriers for index cases and an additional 35 carriers in relatives by genetic sequencing. Thus, the proportion of individuals with FH-causing variants was 68.5% in index cases and 71.0% in all the participants. To put it in more detail, a total of 72 index cases carried known FH-causing variants and only 7 patients did not carry any variants. Of the residual 45 patients who carried novel variants with uncertain significance, we predicted their pathogenicity using bioinformatics tools as described in the previous text and found that 8 patients carried likely pathogenic variants. CNVs were attributed to FH in 5 patients. The prevalence of different genotypes in the present study are listed in Table 2. Of the 85 index cases with FH-causing variants, 54 patients were LDLR heterozygotes, only 1 participant was an APOB heterozygote, and none of them were PCSK9 heterozygotes. Furthermore, 5 double heterozygotes, 12 LDLR compound heterozygotes, and 4 LDLR homozygotes were also identified. Supplemental Table 1 shows the clinical characteristics of FH patients with variants in 2 alleles. Compared with those with compound and double heterozygotes, patients with true homozygotes were younger, were more likely to have xanthoma or corneal arcus, and had significantly higher concentrations of LDL-C and total cholesterol.
Table 2.
Prevalence of Different FH Genotypes
| Index Cases (n = 124) |
Index Cases + Relatives (n = 169) |
|||||
|---|---|---|---|---|---|---|
| n (%) | LDL-C (mmol/l) | Age (yrs) | n (%) | LDL-C (mmol/l) | Age (yrs) | |
| No variant | 39 (31.5) | 5.61 ± 2.39 | 48 ± 12 | 49 (29) | 5.65 ± 2.27 | 46 ± 14 |
| LDLR | ||||||
| Missense | 34 (27.4) | 5.45 ± 2.43 | 45 ± 14 | 54 (32.0) | 5.71 ± 2.07 | 43 ± 17 |
| Nonsense | 10 (8.1) | 5.84 ± 2.05 | 52 ± 12 | 13 (7.7) | 6.38 ± 2.12 | 52 ± 11 |
| Splicing | 2 (1.6) | 5.68 ± 0.37 | 48 ± 11 | 2 (1.2) | 5.68 ± 0.37 | 48 ± 11 |
| Frameshift | 9 (7.3) | 5.58 ± 1.25 | 38 ± 8 | 17 (10.1) | 5.37 ± 1.21 | 41 ± 17 |
| APOB | 1 (0.8) | 3.71 | 46 | 1 (0.6) | 3.71 | 46 |
| Two alleles | ||||||
| Compound HeFH | 12 (9.7) | 5.83 ± 2.02 | 39 ± 16 | 13 (7.7) | 6.06 ± 2.10 | 40 ± 16 |
| Double HeFH | 5 (4.0)∗ | 6.22 ± 2.15 | 44 ± 12 | 7 (4.1) | 6.35 ± 1.79 | 41 ± 13 |
| HoFH | 4 (3.2) | 10.95 ± 2.15 | 31 ± 8 | 4 (2.4) | 10.95 ± 2.15 | 31 ± 8 |
| CNV | 5 (4.0) | 6.52 ± 3.26 | 45 ± 20 | 5 (3.0) | 6.52 ± 3.26 | 45 ± 20 |
| Minor genes | 3 (2.4) | 4.90 ± 2.00 | 32 ± 21 | 4 (2.4) | 5.38 ± 1.89 | 41 ± 25 |
Values are mean ± SD unless otherwise indicated.
APOB = apolipoprotein B; CNV = copy number variants; HeFH = heterozygous familial hypercholesterolemia; HoFH = homozygous familial hypercholesterolemia; LDLR = low-density lipoprotein receptor; other abbreviations as in Table 1.
4 patients with LDLR + PCSK9, 1 patient with LDLR + APOB.
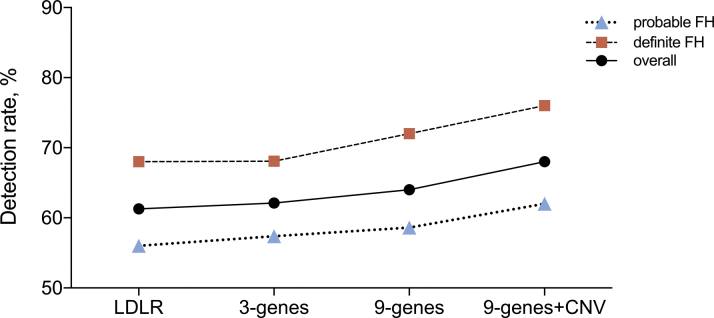
We compared the detection rates in the index cases when different sequencing strategies were applied (Figure 1). The proportion of FH caused by small-scale variants in LDLR, APOB, and PCSK9 genes was 62.1% and then only increased by 6.5% after further analyzing minor FH-related variants and CNV. Further, there was no significant difference with regard to the detection rate between definite and probable FH in different sequencing strategies (all p > 0.05). However, when it came to the detailed genotype of index cases with FH, a significant difference was presented between definite and probable FH (p = 0.012), as shown in Figure 2.
Figure 1.
The Comparison of Detection Rate of FH-Associated Variants in Index Cases According to Different Sequencing Strategies
Three genes refers to LDLR, APOB, and PCSK9. Nine genes refers to LDLR, APOB, PCSK9, LDLARP1, APOE, LIPA, STAP1, and ABCG5/8. ABCG5/8 = ATP-binding cassette transporter family members 5 and 8; APO = apolipoprotein; CNV = copy number variant; FH = familial hypercholesterolemia; LDLR, low-density lipoprotein receptor; LIPA = lipase A; PCSK9 = proprotein convertase subtilisin/kexin type 9. STAP1 = signal transducing adaptive family member 1.
Figure 2.
The Genotypes and Comparison Between Definite FH and Probable FH in Index Cases
HeFH = heterozygous FH; HoFH = homozygous FH; other abbreviations as in Figure 1.
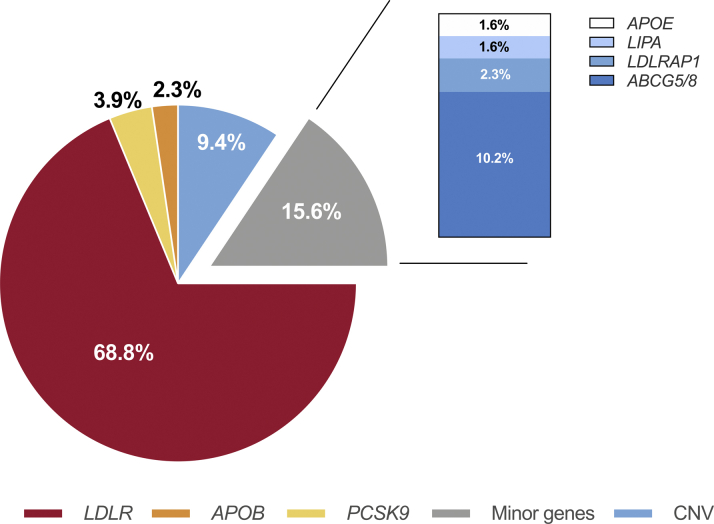
The overall distribution of pathogenic/likely pathogenic FH-related variants in all index cases was shown in Figure 3. In detail, of the total 128 risk variants, the majority was in the LDLR gene, accounting for 68.8% (n = 88). The APOB and PCSK9 variants accounted for 2.3% (3 of 128) and 3.9% (5 of 128), respectively. There were 12 distinct CNVs identified in the current study, with 4 LDLR CNVs; 4 STAP1 CNVs; and 1 CNV each of PCSK9, LIPA, APOE, and ABCG5. All pathogenic/likely pathogenic variants and CNVs found in the current study were summarized in Supplemental Tables 2, 3, and 4. The further family evaluation of the variants was provided in Supplemental Table 5.
Figure 3.
The Distribution of Pathogenic/Likely Pathogenic FH-Causing Variants in Index Cases
Minor genes refers to LDLARP1, APOE, LIPA, STAP1, and ABCG5/8. Abbreviations as in Figure 1.
Finally, we have reviewed the published data and summarized recent publications that also conducted the variant analysis in an expanding manner, including those rare FH-related variants, in Supplemental Table 6. In view of the different participant inclusion, ethics, and diagnostic criteria, the final detection rates varied.
Discussion
In the current study, a total of 169 patients with clinical probable/definite FH received NGS for 9 genes. Overall, we found that FH-associated variants were detected in 85 index cases with a prevalence of 68.5%, and the detection rate increased by 6.5% when LDLRAP1, LIPA, STAP1, APOE, and ABCG5/8 variants and CNVs were included (Central Illustration). To our knowledge, there has been few studies on unconventional genetic screening (minor genes and CNVs) for patients with FH in China.
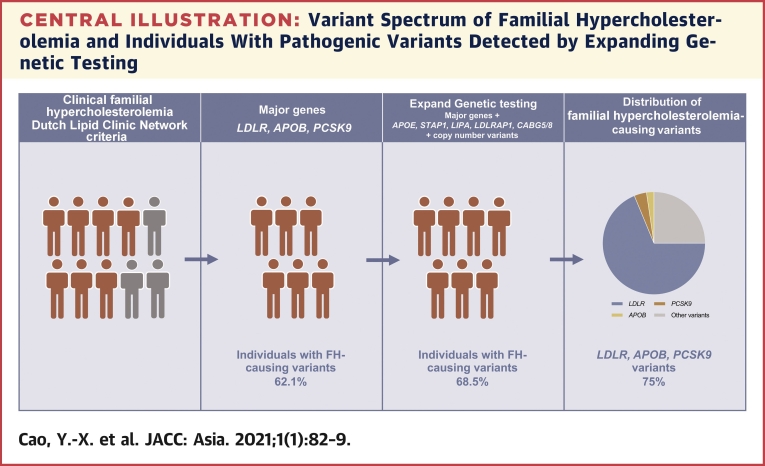
Central Illustration.
Variant Spectrum of Familial Hypercholesterolemia and Individuals With Pathogenic Variants Detected by Expanding Genetic Testing
Expanding genetic testing was performed in patients with clinical familial hypercholesterolemia. The percentage of individuals caused by pathogenic variants in LDLR, APOB, and PCSK9 genes was 62.1%, and the percentage further increased to 68.5% when other genes and copy number variants were included. LDLR, APOB, and PCSK9 variants occupied 75% of all the familial hypercholesterolemia–associated variants. ABCG5/8 = ATP-binding cassette transporter family members 5 and 8; APO = apolipoprotein; LDLR, low-density lipoprotein receptor; LIPA = lipase A; PCSK9 = proprotein convertase subtilisin/kexin type 9; STAP1 = signal transducing adaptive family member 1.
Originally, FH was described as elevated LDL-C due to the loss-function of LDLR (15). With the advancement of DNA sequencing and a deeper understanding of the genetic basis of FH, its pathogenesis has been nearly redefined (16,17). In the present study, we implemented an NGS strategy for sequencing the promoter, coding, and exon-intron boundary regions of 9 FH-related genes as well as CNVs and harbored FH-associated variants in 68.5% of the index cases. The detection rate is influenced by factors including ethnicity, sequencing methodology, clinical diagnostic algorithms, and participant selection. Wang et al. (18) applied target NGS for variants in 9 FH-causing genes and evaluated CNV and polygenic scores in a cohort of 313 patients with LDL-C >5.0 mmol/l. They found that the monogenic detection rate was 47.3% in total and raised to 53.7% when LDLR-CNV were included. When solely focusing on the classical variants in LDLR, APOB, and PCSK9 genes, our study observed FH-causing variants in 62.1% of the participants, broadly consistent with previous studies with a detection rate of approximately 44.6% to 73% in Europe, South America, and Asia (19, 20, 21). Not surprisingly, LDLR gene variants occupied the majority, with a proportion of 68.8% in all FH-causing variants.
Recent publications have revealed that other candidate genes except for LDLR, APOB, and PCSK9 also involved in the manifestation of FH in individuals without indefinable variants in these 3 genes (22). Fouchier et al. (23) first identified 4 variants in STAP1 associated with autosomal-dominant hypercholesterolemia (ADH) using parametric linkage analysis combined with exome sequencing. However, subsequent studies have provided inconsistent evidence for STAP1 as a possible causal gene for FH. Recently, Loaiza et al. (24) analyzed the cosegregation of STAP1 and failed to confirm a major role of STAP1 in the etiology of FH. Thus, to make our results more rigorous, participants with LDLR plus STAP1 variants were not categorized as double heterozygotes in the present study. The APOE gene accounts for a significant fraction of the variation in plasma cholesterol levels and serves a vital role in the lipid metabolism (25). Marduel et al. (26) reported an exceptionally large family including 14 members with ADH and confirmed a variant p.Leu167del in the APOE gene, previously reported to be associated with familial combined hyperlipidemia, also involved in ADH. Furthermore, another phenocopy of FH that has recently taken on increased importance is a rare disorder known as lysosomal acid lipase deficiency due to variants in LIPA, which encodes for lysosomal acid lipase (27). An extremely rare disorder of hypercholesterolemia also exhibits in a recessive mode of inheritance caused by variants in the LDLRAP1 gene (28). The complete loss-of-function of the LDLRAP1 gene resulted in the absence of an adopt protein, which contains a conserved phosphotyrosine-binding domain and functions as an accessory adaptor protein that interacts with the LDLR. Consequently, the LDLR cannot internalize and accumulate at the surface of hepatocytes (15). In parallel, our current genetic sequencing also included ABCG5/8 genes, which have been previously recognized as the cause of sitosterolemia (16). Patients with sitosterolemia also showed heterogeneity in regard of phenotype and parts of individuals had severe hypercholesterolemia with accelerated atherosclerosis and premature CVD, which resemble FH and may cause confusions at initial clinical visiting (29). Thus, the sequencing of ABCG5/8 could assist in the diagnosis and subsequent medical intervention, especially when the measurement of plant sterol is lacking. It is noteworthy that 2 index cases were found carrying variants in ABCG5 with confirmations from their relatives and strengthened the need for its screening. Among the 6 minor genes, variants in APOE exert the effect in a dominant pattern as LDLR, APOB, and PCSK9, whereas 2 mutant alleles act recessively for LDLRAP1, LIPA, and ABCG5/8. Hence, although 15.63% pathogenic/likely pathogenic variants were detected in the study, some of them were heterozygous and could not be responsible for the pathogenesis of FH.
Interestingly, our study also identified novel non-LDLR CNVs, which have been reported for the first time. Iacocca et al. (30) compared the performances of conventional method multiplex ligation-dependent probe amplification (MLPA) and advanced NGS approach in detecting CNVs. They found a 100% concordance in LDLR CNV detection between these 2 methods, suggesting that MLPA can be replaced by NGS to significantly reduce costs, resources, and analysis time. Thus, we applied NGS for CNV detection.
Although the presence of those rare genes has been postulated and discovered, the necessity of extending the sequencing spectrum for patients with FH may be worthy of investigation. On the one hand, sequencing of those rare FH-associated loci could help broaden the range of disease-causing variants and expand our understanding of the complex molecular basis of FH. But, on the other hand, its underlying sense in clinical practice is not always definitive. First, those novel variants often lack evidence of functional assay, and their clinical significances are uncertain. In the current study, we also applied family evaluation for further confirmation of the pathogenicity of the novel variants. Second, the prevalence of variants in those genes is relatively much lower (1). In fact, the detection rate only increased by 6.5% with adding minor genes as well as CNVs in the present study. Last but not least, the cost-effectiveness of NGS should be considered, as the price of sequencing 9 genes is nearly 3 times of that for 3 genes in China. Comprehensively, there may be a reluctance to expand genetic testing for all suspected patients with FH as an initial screening. However, an expanded screening may aid patients negative for conventional variants in LDLR, APOB, and PCSK9 and their relatives.
Study limitations
First, this single-center study with a relatively small sample size could not fully reflect the precise genetic data of Chinese patients with FH. However, FH is underdiagnosed and undertreated, with <1% of patients diagnosed in most countries (2). Thus, it was difficult to enroll a large sample size. Second, about 30% of participants in the cohort were still variant-negative. Recent studies indicated that polygenic hypercholesterolemia due to cumulative effect of LDL-C raising SNPs could explain the phenotype of severe hypercholesterolemia (15). The current study did not evaluate the polygenic scores, considering that the best risk score model and its threshold for diagnosis was not clear in Chinese population. Alternatively, we will address this issue in the further study. Third, the families screened were limited with only 1 or 2 relatives included, which may possibly be due to the 1-child policy in China (31).
Conclusions
The current study replenishes the knowledge of variant spectrum of FH in China through the expanding genetic analysis. Major genes should be tested in the initial genetic screening, although the variants in minor genes also could explain phenotypic FH even with a quite low percentage. An expanding clinical genetic testing strategy may be considered for disease-definite diagnosis and management, especially for those patients who are negative for LDLR, APOB, and PCSK9.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE:LDLR, APOB, and PCSK9 genes should be tested in the initial genetic screening, although expanding genetic testing for the variants in minor genes and CNVs also could explain phenotypic FH for those patients who are negative for these 3 genes.
TRANSLATIONAL OUTLOOK: Additional studies should evaluate the diagnostic value of the expanding genetic testing for FH.
Funding Support and Author Disclosures
This work was supported by the Capital Health Development Fund (201614035), CAMS Major Collaborative Innovation Project (2016-I2M-1-011). The authors have reported that theyhave no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank all of the staff and participants of this study for their important contributions.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental tables, please see the online version of this paper.
Appendix
References
- 1.Wilemon K.A., Patel J., Aguilar-Salinas C., et al. Reducing the clinical and public health burden of familial hypercholesterolemia: a global call to action. JAMA Cardiol. 2020;5:217–229. doi: 10.1001/jamacardio.2019.5173. [DOI] [PubMed] [Google Scholar]
- 2.Nordestgaard B.G., Chapman M.J., Humphries S.E., et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013;34:3478–3490. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuchel M., Bruckert E., Ginsberg H., et al. Homozygous familial hypercholesterolaemia: new insights and guidance for clinicians to improve detection and clinical management. A position paper from the Consensus Panel on Familial Hypercholesterolaemia of the European Atherosclerosis Society. Eur Heart J. 2014;35:2146–2157. doi: 10.1093/eurheartj/ehu274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gidding S.S., Champagne M.A., de Ferranti S.D., et al. The agenda for familial hypercholesterolemia: a scientific statement from the American Heart Association. Circulation. 2015;132:2167–2192. doi: 10.1161/CIR.0000000000000297. [DOI] [PubMed] [Google Scholar]
- 5.Sturm A., Knowles J., Gidding S., et al. Clinical genetic testing for familial hypercholesterolemia: JACC Scientific Expert Panel. J Am Coll Cardiol. 2018;72:662–680. doi: 10.1016/j.jacc.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 6.Leigh S., Futema M., Whittall R., et al. The UCL low-density lipoprotein receptor gene variant database: pathogenicity update. J Med Genet. 2017;54:217–223. doi: 10.1136/jmedgenet-2016-104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amor-Salamanca A., Castillo S., Gonzalez-Vioque E., et al. Genetically confirmed familial hypercholesterolemia in patients with acute coronary syndrome. J Am Coll Cardiol. 2017;70:1732–1740. doi: 10.1016/j.jacc.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Di Taranto M.D., Giacobbe C., Fortunato G. Familial hypercholesterolemia: a complex genetic disease with variable phenotypes. Eur J Med Genet. 2020;63:103831. doi: 10.1016/j.ejmg.2019.103831. [DOI] [PubMed] [Google Scholar]
- 9.Brinton E.A., Hopkins P.N., Hegele R.A., et al. The association between hypercholesterolemia and sitosterolemia, and report of a sitosterolemia kindred. J Clin Lipidol. 2018;12:152–161. doi: 10.1016/j.jacl.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 10.Berberich A.J., Hegele R.A. The complex molecular genetics of familial hypercholesterolaemia. Nat Rev Cardiol. 2019;16:9–20. doi: 10.1038/s41569-018-0052-6. [DOI] [PubMed] [Google Scholar]
- 11.Weng S., Kai J., Akyea R., Qureshi N. Detection of familial hypercholesterolaemia: external validation of the FAMCAT clinical case-finding algorithm to identify patients in primary care. Lancet Public Health. 2019;4:256–264. doi: 10.1016/S2468-2667(19)30061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J.J., Li S., Zhu C.G., et al. Familial hypercholesterolemia phenotype in Chinese patients undergoing coronary angiography. Arterioscler Thromb Vasc Biol. 2017;37:570–579. doi: 10.1161/ATVBAHA.116.308456. [DOI] [PubMed] [Google Scholar]
- 13.Ruel I., Aljenedil S., Sadri I., et al. Imputation of baseline LDL cholesterol concentration in patients with familial hypercholesterolemia on statins or ezetimibe. Clin Chem. 2018;64:355–362. doi: 10.1373/clinchem.2017.279422. [DOI] [PubMed] [Google Scholar]
- 14.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–12. [Google Scholar]
- 15.Mariano C., Alves A.C., Medeiros A.M., et al. The familial hypercholesterolaemia phenotype: Monogenic familial hypercholesterolaemia, polygenic hypercholesterolaemia and other causes. Clin Genet. 2020;97:457–466. doi: 10.1111/cge.13697. [DOI] [PubMed] [Google Scholar]
- 16.Reeskamp L.F., Volta A., Zuurbier L., et al. ABCG5 and ABCG8 genetic variants in familial hypercholesterolemia. J Clin Lipidol. 2020;14:207–217. doi: 10.1016/j.jacl.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Huang C.C., Charng M.J. Genetic diagnosis of familial hypercholesterolemia in Asia. Front Genet. 2020;11:833. doi: 10.3389/fgene.2020.00833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Dron J.S., Ban M.R., et al. Polygenic versus monogenic causes of hypercholesterolemia ascertained clinically. Arterioscler Thromb Vasc Biol. 2016;36:2439–2445. doi: 10.1161/ATVBAHA.116.308027. [DOI] [PubMed] [Google Scholar]
- 19.Jannes C., Santos R., de Souza Silva P., et al. Familial hypercholesterolemia in Brazil: cascade screening program, clinical and genetic aspects. Atherosclerosis. 2015;238:101–107. doi: 10.1016/j.atherosclerosis.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Grenkowitz T., Kassner U., Wühle–Demuth M., et al. Clinical characterization and mutation spectrum of German patients with familial hypercholesterolemia. Atherosclerosis. 2016;253:88–93. doi: 10.1016/j.atherosclerosis.2016.08.037. [DOI] [PubMed] [Google Scholar]
- 21.Fairoozy R.H., Futema M., Vakili R., et al. The genetic spectrum of familial hypercholesterolemia (FH) in the Iranian population. Sci Rep. 2017;7:17087. doi: 10.1038/s41598-017-17181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Futema M., Plagnol V., Li K., et al. Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J Med Genet. 2014;51:537–544. doi: 10.1136/jmedgenet-2014-102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouchier S., Dallinga-Thie G., Meijers J., et al. Mutations in STAP1 are associated with autosomal dominant hypercholesterolemia. Circ Res. 2014;115:552–555. doi: 10.1161/CIRCRESAHA.115.304660. [DOI] [PubMed] [Google Scholar]
- 24.Loaiza N., Hartgers M.L., Reeskamp L.F., et al. Taking one step back in familial hypercholesterolemia: STAP1 does not alter plasma LDL (low-density lipoprotein) cholesterol in mice and humans. Arterioscler Thromb Vasc Biol. 2020;40:973–985. doi: 10.1161/ATVBAHA.119.313470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awan Z., Choi H.Y., Stitziel N., et al. APOE p.Leu167del mutation in familial hypercholesterolemia. Atherosclerosis. 2013;231:218–222. doi: 10.1016/j.atherosclerosis.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Marduel M., Ouguerram K., Serre V., et al. Description of a large family with autosomal dominant hypercholesterolemia associated with the APOE p.Leu167del mutation. Hum Mutat. 2013;34:83–87. doi: 10.1002/humu.22215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikhailova S., Ivanoshchuk D., Timoshchenko O., Shakhtshneider E. Genes potentially associated with familial hypercholesterolemia. Biomolecules. 2019;9:807. doi: 10.3390/biom9120807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spina R., Noto D., Barbagallo C.M., et al. Genetic epidemiology of autosomal recessive hypercholesterolemia in Sicily: identification by next-generation sequencing of a new kindred. J Clin Lipidol. 2018;12:145–151. doi: 10.1016/j.jacl.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Kawamura R., Saiki H., Tada H., Hata A. Acute myocardial infarction in a 25-year-old woman with sitosterolemia. J Clin Lipidol. 2018;12:246–249. doi: 10.1016/j.jacl.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Iacocca M., Wang J., Dron J., et al. Use of next-generation sequencing to detect gene copy number variation in familial hypercholesterolemia. J Lipid Res. 2017;58:2202–2209. doi: 10.1194/jlr.D079301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu X., Pang J., Wang X., et al. Reverse cascade screening for familial hypercholesterolemia in high-risk Chinese families. Clin Cardiol. 2017;40:1169–1173. doi: 10.1002/clc.22809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.