Fig. 4. Discovery of residues within compound binding pocket that define susceptibility to VGTI-A3–03.
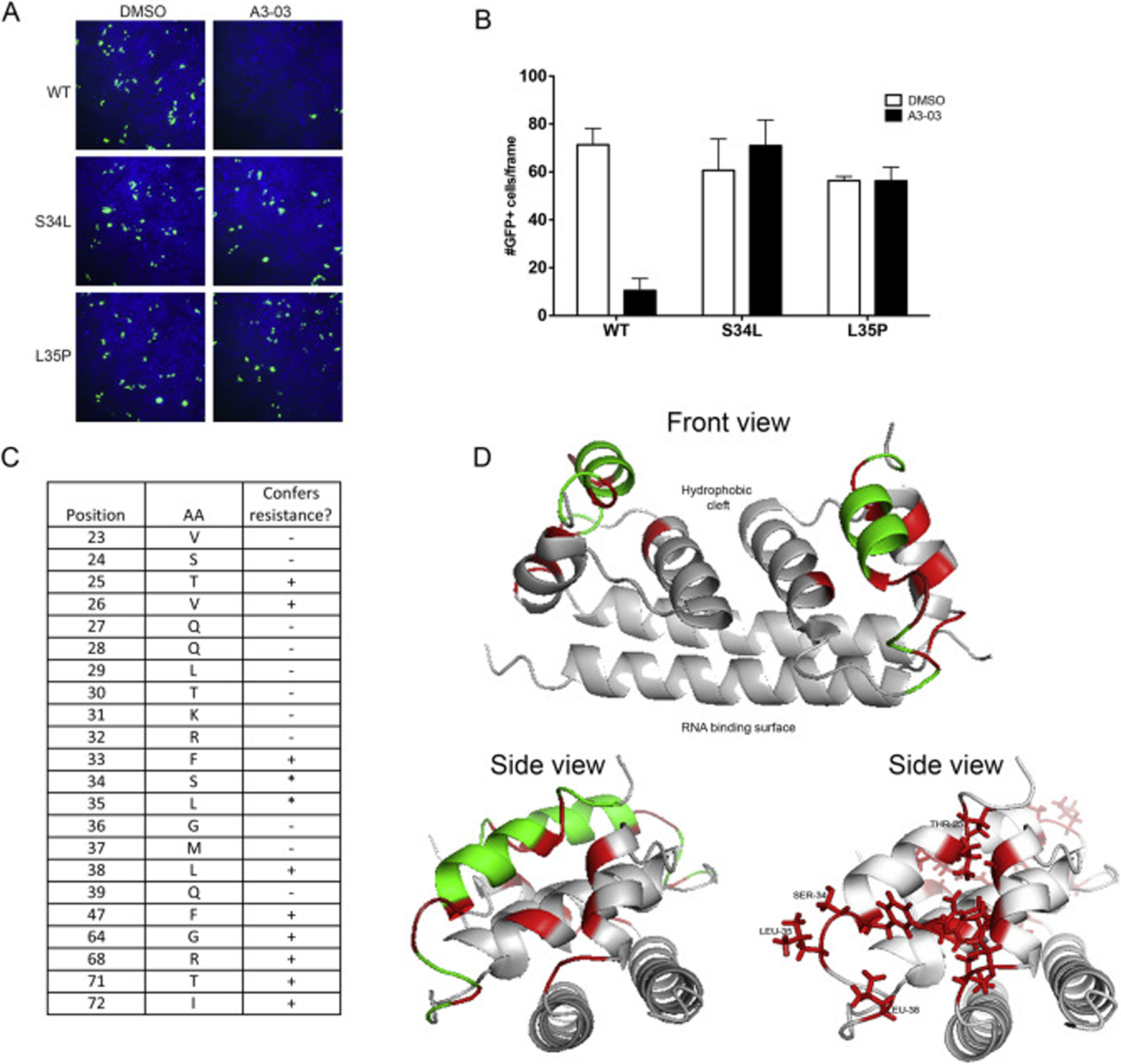
(A) DENV2 reporter virus particles (RVPs) expressing GFP were produced by transfection of DRep2A-expressing cells with a plasmid encoding DENV2 structural proteins (wild type or containing the C S34L or L35P mutations). At 1 day p.t., media was removed and replaced with 1uM VGTI-A3–03 or DMSO control. Supernatants containing RVPs were collected at 4 days p.t., purified over a sorbitol cushion, and used to infect naïve HEK293 cells at MOI = 5 IU/ml. GFP + cells were assayed at 3 days p.i. (B) GFP + cells were quantitated from 6 independent frames using ImageJ software. (C) RVPs constructed with indicated C residues mutated to alanines were tested for resistance conference to A3–03 as above. Residues that did not tolerate mutation to alanine, as indicated by no RVP production, are indicated with an asterisk. (D) Front and side views of MacPyMol depiction of the C dimer (PBD: 1R6R) with resistance conferring C mutations (red) or resistance neutral (green). The hydrophobic cleft, where the ER membrane is predicted to interact, and the putative RNA binding surface are indicated on front view. Data are presented as mean ± SEM in triplicate. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
