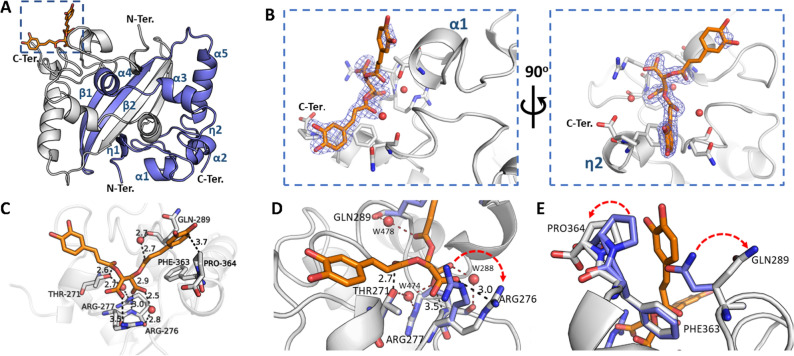
Figure 4.
The crystal structure of SARS-CoV-2 N protein CTD binding chicoric acid (CA) reveals a network of polar contacts and structural readjustments to accommodate the symmetric ligand in the CA-N protein binding site. (A) Cartoon representation of the N protein CTD dimer crystal structure depicting its secondary structure elements (in blue), including two 310 (η) helices, five α-helices and two antiparallel β-strands and the CA binding site (inset highlighted by the blue dashed square). (B) Detailed CA-binding site from the inset of panel (A). The CA molecule is represented as sticks (orange) with its electron-density map in blue. CA binds to a shallow pocket formed between α-helices 1–2 and η-helix 2, close to the C-terminus (C-Ter). (C) CA atomic interactions with the N protein residues. The CA carboxyl groups are at ideal distances to engage electrostatic interactions and hydrogen bonds with Arg276 side chain (NH1 atom), Arg277 main chain amine and a structural water molecule (W288) stabilized by Arg276 NH2. Thr271 and Gln289 can further position hydrogen bond donors (Thr271Oγ and a structural water molecule, W478, stabilized by the Gln289 carbonyl) to engage a symmetric interaction with the carbonyl groups from both caffeoyl units of CA. One of the catechol motifs of CA is well accommodated near the C-terminal Pro364, showing a well-defined electron density (vide panel B). (D,E) Superposition of the CA-binding site in the native N protein CTD (blue sticks, PDB ID 7UXX) and CA-N protein CTD complex (grey sticks (PDB ID 7UXZ) highlighting structural readjustments induced by CA binding (highlighted by red arrows). Figures were generated with Pymol (Schroedinger Inc.). Polar contacts are indicated by dashed lines with the measured distances in Angstroms. Oxygen atoms are shown in red, nitrogen atoms in blue. Water molecules are represented as red spheres.