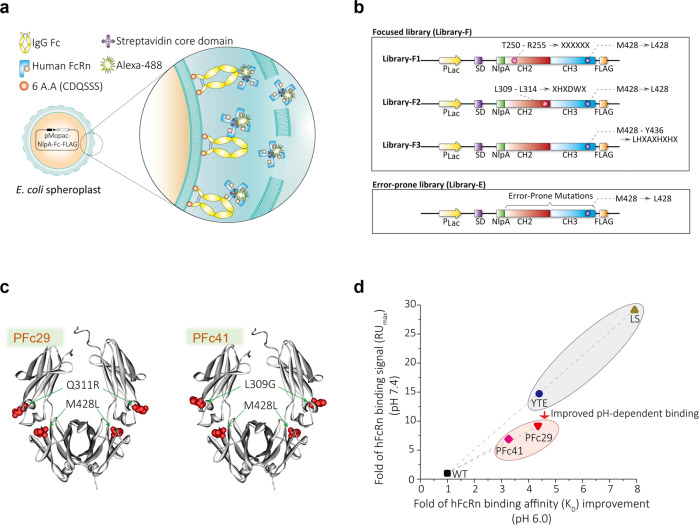
Fig. 1. Isolation of Fc variants with improved FcRn binding.
a Screening strategy for isolating Fc variants using the bacterial display. b Schematic diagram displaying the expression cassettes of the mutagenized Fc libraries (Library-F and Library-E). c Solid ribbon structure showing the mutation sites for the isolated Fc variants. The mutations identified for PFc29 (left) and PFc41 (right) are overlaid on the structure of the Fc region of the human IgG crystal structure (PDB: 1HZH). d The dot plot shows the improved hFcRn binding of the Fc variants relative to a wild-type Fc at pH 6.0 and pH 7.4. The x-axis and y-axis of the dot plot indicate the fold improvement of hFcRn binding affinity (KD) for the Fc variants at pH 6.0 and the fold improvement of hFcRn binding signal (RUmax) for the Fc variants at pH 7.4 relative to a wild-type Fc, respectively. The Fc variants and wild-type Fc were introduced into trastuzumab, and KD and RUmax were measured using an SPR analysis.