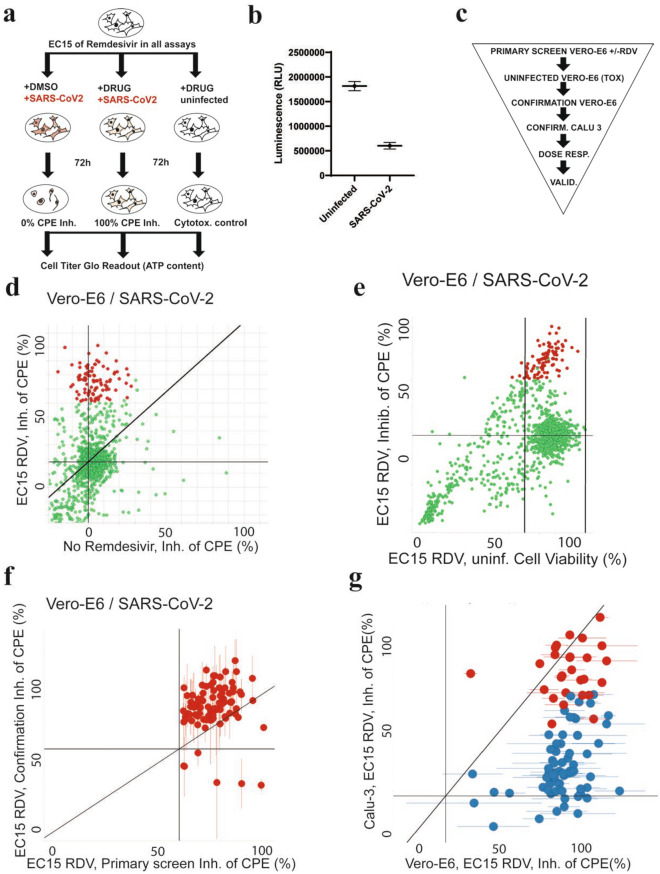
Figure 1.
Primary screening results identifying compounds increasing antiviral effects of remdesivir (RDV). (a) Assay outline: Vero-E6 cells are added to 384 well plates, treated with DMSO (left panel) or drug (middle panel), infected with SARS-CoV-2 and incubated for 72 h to observe cytopathic effect (CPE; left panel). Effective drug treatment inhibits occurrence of CPE (middle panel). CPE is measured by quantifying ATP content in viable cells using a luminescent assay (Cell-Titer Glo). The right panel shows the cytotoxicity control, treating cells with drugs but without virus. (b) Screening assay performance. Average Luminescence is shown for the Vero E6 primary screen in presence of EC15 of remdesivir (n = 144, 24 wells each from 6 screening plates), error bars indicate standard deviation. “Uninfected”: positive control (equivalent to 100% inhibition of CPE), “SARS-CoV-2”: negative control, infected and treated with DMSO (equivalent to 0% inhibition of CPE). Z’ = 0.63 ± 0.04. (c) Screening paradigm outline in presence of EC15 of remdesivir. Cells infected with SARS-CoV-2 unless indicated. Dose resp.—Dose response; Valid.—Validation in orthogonal assays. (d) Primary screen results for 1200 approved drugs tested in Vero E6 cells infected with SARS-CoV-2 in the absence (x-axis) and presence (y-axis) of EC15 of remdesivir. Inhibition of CPE (%) is shown. The horizontal line indicates the background activity of EC15 of remdesivir (not subtracted). Diagonal line: 1:1 correlation. Red: high priority hits with a cutoff of > 60% inhibition of CPE in presence of remdesivir. (e) As in (d), but with cell viability data from cytotoxicity control (uninfected) on the x-axis. Vertical line: Cell viability of 70%. (f) Confirmation of > 95% of high priority hits from (e) after compound cherrypicking; assay conditions as in (e), Vero-E6 cells infected with SARS-CoV-2 in presence of EC15 of remdesivir; x-axis indicates primary screening results (Inhibition of CPE, %), y-axis confirmation results (Inhibition of CPE, %). Horizontal and vertical lines indicate hit progression cutoff from primary screen, diagonal line 1:1 correlation. Error bars indicate standard deviation. (g) 26 compounds (red; labeled) are active in both Vero E6 and human lung epithelial Calu-3 cells infected with SARS-CoV-2 in presence of EC15 of remdesivir. Inhibition of CPE (%) is shown on the x-axis for Vero E6, on the y-axis for Calu-3 cells.