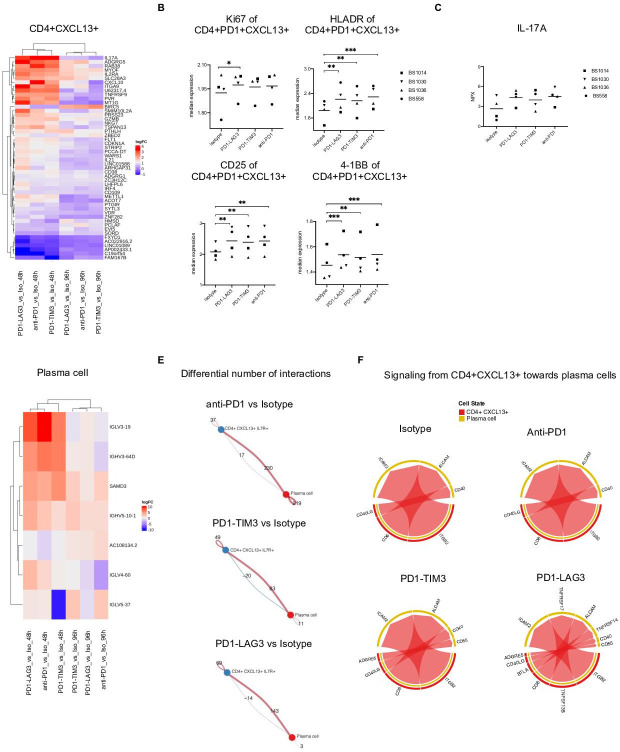
Figure 5.
Transcriptional changes are induced by single or dual checkpoint blockade within CD4+CXCL13+ cells which influence interaction with plasma cells. (A) Top differentially expressed genes showing strongest change among all contrasts (average logCPM >0, adjusted p value <0.05) in the CD4+CXCL13+ cluster. (B) Median marker intensity of Ki67, HLA-DR, 4-1BB and CD25 on the CD4+CXCL13+ population in the four responsive tumor suspension in different treatment conditions. Clustering was conducted using FlowSOM (*p<0.05, **p<0.01, ***p<0.001, one-way ANOVA with multiple comparisons). (C) of IL-17A measured by Olink technology in the supernatant of the four responsive tumor suspension across different treatment versus control comparisons (FDR <0.05, comparing each treatment with control isotype). (D) Top differentially expressed genes showing strongest change among all contrasts (average logCPM >0, adjusted p value <0.05) in the plasma cell cluster. (E) Circle plots showing the differential number of interactions between the CD4+CXCL13+ and the plasma cell clusters, in each treatment compared with control, at 96 hours. Calculated using Cellchat.39 (F) Chord diagram of ligand-receptor pairs were mediating the communications from CD4+CXCL13+ toward plasma cells, in each treatment or control condition, at 96 hours, calculated using Cellchat.39 ANOVA, analysis of variance; CPM, counts per million; FDR, false discovery rate; IL-17A, interleukin-17A; LAG-3, lymphocyte-activation gene 3; PD-1, programmed cell death protein 1; TIM-3, T cell immunoglobulin and mucin-domain containing-3.